Learninsta presents the core concepts of Microbiology with high-quality research papers and topical review articles.
Microbial Genetics Transcription
An important feature of RNA synthesis is that even though the DNA molecule being copied is double stranded, in any particular region of DNA only one strand serves as a template. The DNA strand copied into RNA molecule is called CODING OR SENSE STRAND. The synthesis of RNA consists of five discrete stage (Figure 12.2):
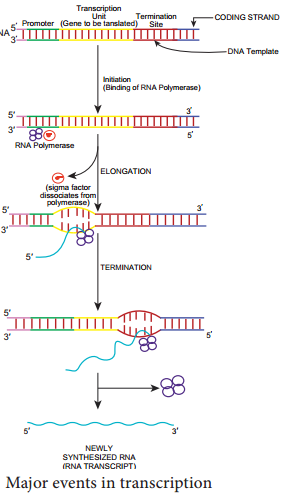
1. Promoter recognition:
RNA polymerase binds to DNA within a specific base sequence (20-200 bases long) called a promoter. The sequence TATAAT (or a nearly identical sequence) often called a pribnow box or – 10 region is found as part of all prokaryotic promoters.
The RNA polymerase of the bacterium E.coli consists of five protein subunits. Four of the subunits comprise the core enzyme (catalyzes the joining of the nucleoside triphosphates to the RNA) and fifth subunit, the σ subunit (required for promoter binding).
2. Local unwinding
Local unwinding of DNA occurs and RNA polymerase forms an open promoter complex.
3. Nucleoside Triphosphate
The first nucleoside triphosphate is placed at polymerization start site (near to the initial binding site) and synthesis begins.
4. RNA
RNA polymerase then moves along the DNA, adding ribonucleotides, to the growing RNA chain.
5. RNA polymerase
RNA polymerase reaches chain termination sequence and both the newly synthesized RNA and the polymerase are released. Two kinds of termination events are known those that are self – terminating (dependent on the base sequence only) and those that require the presence of the termination protein Rho.
Initiation of a second round of transcription need not await completion of the first, for the promoter becomes available once RNA polymerase has polymerized 50-60 nucleotides. In bacteria most mRNA molecules are degraded within a few minutes after synthesis. This degradation enables cells to dispense with molecules
that are no longer needed.
In prokaryotes mRNA molecules commonly contain information for the amino acid sequences of several different polypeptide chains. In this case, such a molecule is called polycistronic mRNA. Cistron is a term used to mean a base sequence encoding a single polypeptide chain.
The genes contained in polycistronic mRNA molecule (Figure 12.3) often encode the different portions of a metabolic pathway. For example, in E. coli the ten enzymes needed to synthesize histidine are encoded in one mRNA molecule.
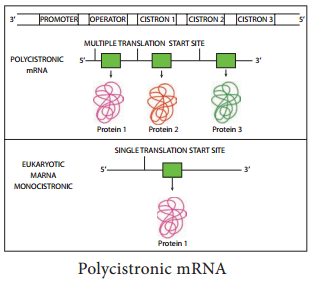
In prokaryotes the immediate product of transcription (called the primary transcript) is mRNA, in contrast in eukaryotes the primary transcript must be converted to mRNA. This conversion called RNA processing consists of two types of events – modification of termini and excision of untranslated sequences (noncoding sequence or introns) embedded within coding sequences (exons).
Introns excision and the joining of exons to form an mRNA molecule is called RNA splicing. The introns are present in almost all eukaryotic transcripts but are rare in the free – living unicellular eukaryotes such as yeast. Some bacterial genes do contain introns.
Synthesis of rRNA and tRNA Ribosomal RNA and tRNA are also transcribed from genes. The production of these molecules is not as direct as synthesis of bacterial mRNA. The main difference is that these RNA molecules are excised from large primary transcripts. Highly specific RNA excise rRNA and tRNA from these large transcripts, and other enzymes produce the modified bases in tRNA.