NCERT Exemplar Solutions for Class 12 Biology chapter 6 Molecular Basis of Inheritance
These Solutions are part of NCERT Exemplar Solutions for Class 12 Biology. Here we have given NCERT Exemplar Solutions for Class 12 Biology chapter 5 Principles of Inheritance and Variation
Multiple Choice Questions
Question 1.
In a DNA strand, the nucleotides are linked together by
(a) glycosidic bonds
(b) phosphodiester bonds
(c) peptide bonds
(d) hydrogen bonds.
Answer:
(b) : A nucleotide has three components – nitrogenous base, a pentose sugar (ribose in case of RNA and deoxyribose for DNA) and a phosphate group. There are two types of nitrogenous bases – purines (adenine and guanine) and pyrimidines (cytosine, thymine and uracil). A nitrogenous base is linked to the pentose sugar through a N-glycosidic linkage to form a nucleoside. When a phosphate group is linked to 5′ -OH of a nucleoside through phosphoester linkage, a correspondingnucleotide is formed. The nucleotides are linked through 3′ – 5′ phosphodiester linkage to form a dinucleotide.
Question 2.
A nucleoside differs from a nucleotide. It lacks the
(a) base
(b) sugar
(c) phosphate group
(d) hydroxyl group.
Answer:
(c) : A purine or pyrimidine base joined with a pentose sugar, either ribose or deoxyribose, is a nucleoside. A nucleotide is a nucleoside with one or more phosphate groups attached to the sugar.
Question 3.
Both deoxyribose and ribose belong to a class of sugars called
(a) trioses
(b) hexoses
(c) pentoses
(d) polysaccharides.
Answer:
(c) : Pentose sugar is a sugar molecule with five carbon ring structure. Both ribose and deoxyribose are pentoses, the latter is formed by deoxygenation of the former at one carbon atom.
Question 4.
The fact that a purine base always paired through hydrogen bonds with a pyrimidine base leads to, in the DNA double helix
(a) the antiparallel nature
(b) the semi-conservative nature
(c) uniform width throughout DNA
(d) uniform length in all DNA.
Answer:
(c) : In DNA chain, the base pairs on the two strands are complementary and a specific purine pairs with a specific pyrimidine. This makes the two chains uniformly (2nm) thick. A larger sized purine lies opposite to the smaller-sized pyrimidine i.e., A pairs with T and C pairs with G giving uniform width throughout DNA.
Question 5.
The net electric charge on DNA and histones is
(a) both positive
(b) both negative
(c) negative and positive, respectively
(d) Zero
Answer:
(c) : DNA is much more organised in eukaryotic chromatin and is associated with a variety of proteins, most prominent of which are histones. Histones are rich in the basic amino acid residues lysines and arginines. Both these amino acid residues carry positive charges in their side chains. Thus, histones are positively charged. Histones are organised to form a unit of eight molecules called as histone octamer. The negatively charged DNA is wrapped around the positively charged histone octamer to form a structure called nucleosome.
Question 6.
The promoter site and the terminator site for transcription are located at
(a) 3′ (downstream) end and 5′ (upstream) end, respectively of the transcription unit
(b) 5′ (upstream) end and 3′ (downstream) end, respectively of the transcription unit
(c) the 5′(upstream) end
(d) the 3′ (downstream) end.
Answer:
(b)
Question 7.
Which of the following statements is the most appropriate for sickle cell anaemia?
(a) It cannot be treated with iron supplements.
(b) It is a molecular disease.
(c) It confers resistance to acquiring malaria.
(d) All of the above
Answer:
(d) : Sickle cell anaemia is an autosomal hereditary disorder in which the erythrocytes become sickle shaped under oxygen deficiency, such as during strenuous exercise and at high altitudes. The disorder or disease is caused by the formation of an abnormal haemoglobin molecule called haemoglobin-S. Sickle cell trait protects against malaria. Several studies have suggested that, sickle haemoglobin might get in the way of Plasmodium parasite infecting RBCs, reducing the number of parasites that actually infect the host cell and thus confer some protection against the disease.
Question 8.
One of the following is true with respect to AUG:
(a) It codes for methionine only.
(b) It is also an initiation codon.
(c) It codes for methionine in both prokaryotes and eukaryotes.
(d) All of the above
Answer:
(d) AUG has dual functions : it acts as initiation codon and also codes for methionine (both in prokaryotes and eukaryotes, although formylated in prokaryotes).
Question 9.
The first genetic material could be
(a) protein
(b) carbohydrates
(c) DNA
(d) RNA.
Answer:
(d) : The first genetic material could be RNA. Evidences suggest that, metabolism, splicing and translation all have evolved around RNA. The first biocatalysts were also RNAs. Even now, some enzymes are made of RNAs, e.g., ribozyme. Gradually during evolution, genetic storage function of RNA was taken over by DNA as RNA being single stranded was more reactive, hence unstable. DNA being double stranded with complementary strands is not only more stable but also resists changes as it has evdlved a process of repair. For biocatalysis, RNA was replaced by protein enzymes. The latter were more stable, efficient and occur in many forms.
Question 10.
With regard to mature mRNA in eukaryotes
(a) exons and introns do not appear in the mature RNA
(b) exons appear but introns do not appear in the mature RNA
(c) introns appear but exons do not appear in the mature RNA
(d) both exons and introns appear in the mature RNA.
Answer:
(b) : Eukaryotic transcripts possess extra segments called introns or intervening sequences or noncoding sequences. They do not appear in mature or processed RNA because post transcriptional processing of the transcript includes splicing i.e., the’ process of removal of introns and fusion of exons to form functional RNAs.
Question 11.
The human chromosome with the highest and least number of genes in them are respectively
(a) chromosome 21 and Y
(b) chromosome 1 and X
(c) chromosome 1 and Y
(d) chromosome X and Y.
Answer:
(c) : Chromosome 1 has 2968 genes while Y-chromosome has 231 genes which respectively are the maximum and minimum number of genes present in a chromosome.
Question 12.
Who amongst the following scientists had no contribution in the development of the double helix model for the structure of DNA?
(a) Rosalind Franklin
(b) Maurice Wilkins
(c) Erwin Chargaff
(d) Meselson and Stahl
Answer:
(d)
Question 13.
DNA isa polymer of nucleotides whicharelinked to each other by 3′-5’phosphodiester bond. To prevent polymerisation of nucleotides, which of the following modifications would you choose?
(a) Replace purine with pyrimidines.
(b) Remove/Replace 3′ OH group in deoxyribose.
(c) Remove/Replace 2′ OH group with some other group in deoxyribose.
(d) Both ‘b’ and ‘c’.
Answer:
(b) : If 3′ OH group is removed/replaced in deoxyribose, there will be no formation of phosphodiester bonds which is formed between 3′ hydroxyl (OH) group of one nucleotide and 5′ phosphate of the other, and hence polymerisation of nucleotides will be prevented.
Question 14.
Discontinuous synthesis of DNA occurs in one strand, because
(a) DNA molecule being synthesised is very long
(b) DNA dependent DNA polymearse catalyses polymerisation only in one direction (5′->3’)
(c) it is a more efficient process
(d) DNA ligase has to have a role.
Answer:
(b) :The DNA-dependent DNA polymerases catalyse polymerisation only in one direction, that is 5′ —> 3′. Consequently, on one strand (the template with polarity 3′ —> 5′), the replication is continuous in 5′ —» 3′ direction, while on the other (the template with polarity 5′ —» 3′), it is discontinuous. A RNA primer is added the 3′ end of the opened region of 5′ —» 3′ strand and on this primer, a short stretch of DNA is synthesised in 5′ —> 3′ direction. Then again, when further uncoiling occurs then a new primer is added at 3′ end which is elongated, and the process continues. These discontinuously synthesised fragments are later joined by the enzyme DNA ligase.
Question 15.
Which of the following steps in transcription is catalysed by RNA polymerase?
(a) Initiation
(b) Elongation
(c) Termination
(d) All of the above
Answer:
(d) : There is single DNA-dependent RN Apoly merase that catalyses transcription of all types of RNA in bacteria. RNA polymerase binds to promoter and initiates transcription (initiation). It uses nucleoside triphosphates as substrate and polymerises them in a template dependent fashion following the rule of complementarity. It somehow also facilitates opening of the helix and continues elongation. Only a short stretch of the prepared RNA remains bound to the enzyme.Once the polymerase reaches the terminator region, the nascent RNA falls off, so also the RNA polymerase. This results in termination of transcription. The RNA polymerase alone is only capable to catalyse the process of elongation. It associates transiently with initiation factor(o) and termination factor(р) to initiate and terminate the transcription respectively. Association with these factors alters the specificity of the RNA polymerase to either initiate or terminate. In eukaryotes, several initiation factors and termination factors along with three types of RNA polymerase each polymerising a specific RNA perform these functions.
Question 16.
Control of gene expression takes place at the level of
(a) DNA replication
(b) transcription
(c) translation
(d) none of the above
Answer:
(b & c) : Regulation of gene expression refers to a very broad term that may occur at various levels. In eukaryotes, the gene expression can be regulated at transcriptional level, post transcriptional processing level, during transport of mRNA from nucleus to the cytoplasm, and at translational level whereas in prokaryotes, control of the rate of transcriptional initiation is the predominant site for control of gene expression.
Question 17.
Regulatory proteins are the accessory proteins that interact with RNA polymerase and affect its role in transcription. Which of the following statements is correct about regulatory proteins?
(a) They only increase expression.
(b) They only decrease expression.
(c) They interact with RNA polymerase but do not affect the expression.
(d) They can act both as activators and as repressors.
Answer:
(d) : In a transcription unit, the activity of RNA polymerase at a given promoter is regulated by interaction with accessory proteins, which affect its ability to recognise start sites. These regulatory proteins can act both positively (activators) and negatively (repressors).
Question 18.
Which was the last human chromosome to be completely sequenced:
(a) Chromosome 1
(b) Chromosome 11
(c) Chromosome 21
(d) Chromosome X
Answer:
(a) : The sequencing of chromosome 1 was completed in May 2006. This was the last of the 24 human chromosomes (22 autosomes and X and Y) to be sequenced.
Question 19.
Which of the following are the functions of RNA?
(a) It is a carrier of genetic information from DNA to ribosomes synthesising polypeptides.
(b) It carries amino acids to ribosomes.
(c) It is a constituent component of ribosomes.
(d) All of the above.
Answer:
(d) : mRNA carries genetic information from DNA to ribosomes for synthesising polypeptide chains. fRNA carries amino acids to ribosomes attached to mRNA for translation. rRNA, is a vital component of ribosomes.
Question 20.
While analysing the DNA of an organism, a total number of 5386 nucleotides were found out of which the proportion of different bases were: Adenine = 29%, Guanine = 17%, Cytosine = 32%, Thymine = 17%. Considering the Chargaff’s rule it can be concluded that
(a) it is a double stranded circular DNA
(b) it is a single stranded DNA
(c) it is a double stranded linear DNA
(d) no conclusion can be drawn.
Answer:
(b) : It cannot be a double stranded DNA because as per Chargaff’s rule for a dsDNA, the ratios between adenine and thymine, and guanine and cytosine are constant and equal. Hence, it is a ssDNA.
Question 21.
In some viruses, DNA is synthesised by using RNA as template. Such a DNA is called
(a) A-DNA
(b) B-DNA
(c) c DNA
(d) rDNA.
Answer:
(c) : DNA synthesis on RNA template occurs in retroviruses or RNA viruses. They have RNA as their genetic material. When they infect a host cell, they first synthesise DNA on RNA template which is known as cDNA or complementary DNA. This cDNA then continues the process of infection.Regulatory proteins are the accessory proteins that interact with RNA polymerase and affect its role in transcription. Which of the following statements is correct about regulatory.
Question 22.
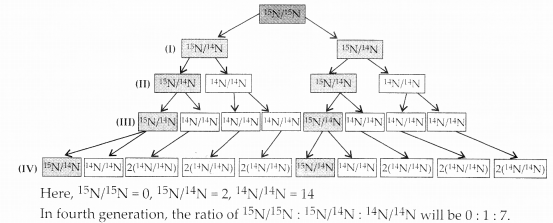
If Meselson and Stahl’s experiment is continued for four generations in bacteria, the ratio of ,5N/15N: 15N/14N: 14N/14N containing DNA in the fourth generation would be
(a) 1:1:0
(b) 1:4:0
(c) 0:1:3
(d) 0:1:7
Answer:
(d) : In Meselson and Stahl’s experiment, parent DNA isolated from E.coli is grown in heavy 15N medium. It was then put in light ,4N medium.Then, when replication occurs the new strand synthesised would have l4N. The process can be represented as follows:
Question 23.
If the sequence of nitrogen bases of the coding strand of DNA in a transcription unit is 5′ – AT G A A T G – 3′, the sequence of bases in its RNA transcript would be
(a) 5′-AUGAAUG-3′
(b) 5′-UACUUAC-3′
(c) 5′-C All UC AU-3′
(d) 5′ – G U A A G U A – 3′.
Answer:
(a) : The sequence of bases in RNA transcript is similar (not complementary) to the coding DNA strand except that in RNA, uracil is present in place of thymine. So, the correct sequence of bases is:
5′ – AUGAAUG – 3′
Question 24.
The RNA polymerase holoenzyme transcribes
(a) the promoter, the structural gene and the terminator region
(b) the promoter and the terminator region
(c) the structural gene and the terminator region
(d) the structural gene only.
Answer:
(d) : Transcription involves three separate processes : initiation, elongation and termination. Initiation begins when the RNA polymerase binds to the promoter which serves only as a target site for binding of the RNA polymerase and is not transcribed. Each gene contains a specific promoter region for guiding the beginning of transcription. This is followed by region of gene (structural gene) that is transcribed and ends with a terminator that stops transcription and is not transcribed.
Question 25.
If the base sequence of a codon in mRNA is 5′-AUG-3′, the sequence of fRNA pairing with it must be
(a) 5′- UAC – 3′
(b) 5′-CAU-3′
(c) 5’-AUG-3′
(d) 5′- GUA – 3′.
Answer:
(b) : The first base of anticodon in 5′ – 3′ direction binds with the third base in codon (reading in 5′ – 3′ direction). Thus, if the base sequence in codon of mRNA is 5′- AUG – 3′, the complementary anticodon will be 3′-UAC – 3′ or 5′- CAl – 3′.
Question 26.
The amino acid attaches to the fRNA at its
(a) 5′-end
(b) 3′-end
(c) Anticodon site
(d) DHU loop.
Answer:
(b) : fRNA has an anticodon loop that has bases complementary to the code, it also has an amino acid acceptor site at which it binds to amino acids. This site lies at the 3′ end opposite the anticodon. fRNAs are specific for each ahrino acid.
Question 27.
To initiate translation, the mRNA first binds to
(a) the smaller ribosomal sub-unit,
(b) the larger ribosomal sub-unit
(c) the whole ribosome
(d) no such specificity exists.
Answer:
(a) : The cellular factory responsible for synthesising proteins is the ribosome. The ribosome consists of structural RNAs and about 80 different proteins. It has two subunits, a large subunit and a small subunit. To start translation, mRNA should bind to small ribosomal unit.
Question 28.
In E. coli, the lac operon gets switched on when
(a) lactose is present and it binds to the repressor
(b) repressor binds to operator
(c) RNA polymerase binds to the operator
(d) lactose is present and it binds to RNA polymerase.
Answer:
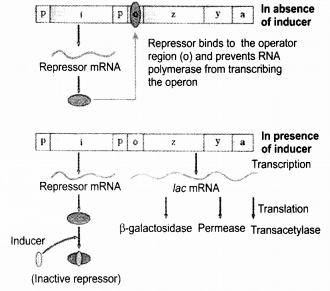
(a) : The repressor of the operon is synthesised (all-the-time-constitutively) from the / gene. The repressor protein binds to the operator region of the operon and prevents RNA polymerase from transcribing the operon. In the presence of an inducer, such as lactose or allolactose, the repressor is inactivated by interaction with the inducer. This allows RNA polymerase access to the promoter and transcription proceeds. Thus, the lac operon gets switched on.
Very Short Answer Type Questions
Question 1.
What is the function of histones in DNA packaging?
Answer:
Maim Histones are positively charged basic proteins which help in condensation of DNA. Negatively charged DNA gets wrapped around positively charged histone octamer to form nucleosome.
Question 2.
Distinguish between heterochromatin and euchromatin. Which of the two is transcriptionally active?
Answer:
Heterochromatin is thicker, densely packed, dark stained part of chromatin. Euchromatin is thinner, loosely packed lightly stained part of chromatin. Transcriptionally active part is euchromatin.
Question 3.
The enzyme DNA polymerase in coli is a DNA dependent polymerase and also has the ability to proofread the DNA strand being synthesised. Explain. Discuss the dual polymerase.
Answer:
DNA polymerase serves as a dual polymerase which serves two functions. It primary function is to add nucleotides according to the template strand in 5′ —> 3′ direction. It simultaneously proof reads the newly formed double strand, as it passes through the polymerase molecule. If the wrong base is inserted, the bond is unstable and spontaneous melting of the newly formed stretch occurs. The new strand gets exposed to the 3′ exonuclease site of the enzyme which removes the mismatched base and some additional nucleotides from the 3’ end. Then, the polymerase activity is continued again. This reduces the chances of error in DNA replication from about one in a million to about one in a hundred million base pairs.
Question 4.
What is the cause of discontinuous synthesis of DNA on one of the parental strands of DNA? What happens to these short stretches of synthesised DNA?
Answer:
New DNA strand is always formed in 5′-3′ direction during DNA replication, over DNA template with 3′-5′ direction. When DNA opens during replication, 3′-5′ strand forms a continuous strand called leading strand. The other template strand with 5′-3′ orientation produces a new short strand over the fork every time when it opens. These short segments are called Okazaki fragments. These segments join together with DNA ligases and form lagging strand.
Question 5.
Given below is the sequence of coding strand of DNA in a transcription unit:
3’AATGCAGCTATTAGG-5′ Write the sequence of
(a) its complementary strand
(b) the mRNA
Answer:
(a) Sequence of complementary strand will be : 5′ -TTACGTCGATAATCC3′
(b) Sequence of mRNA will be : 3′ – A A U G C A G C U A U U A G G 5′
Question 6.
What is DNA polymorphism? Why is it important to study it?
Answer:
Variation at genetic level arisen due to mutations, is called polymorphism. Such variations are unique at particular site of DNA. The polymorphism (variations) in DNA sequences is the basis of genetic mapping and DNA fingerprinting.
Question 7.
Based on your understanding of genetic code, explain the formation of any abnormal haemoglobin molecule. What are the known consequences of such a change?
Answer:
Haemoglobin is made of four polypeptide chains, two a-chains-which are 141 amino acids long and two (3-chains which are 146 amino acids long. In case of sickle cell anaemia, a fault occurs at the sixth amino acid in the (5-chain. The, amino acid should be glutamic acid. In HbS however, it is replaced by valine.

Question 8.
Sometimes cattle or even human beings give birth to their young ones that are having extremely different sets of organs like limbs/ position of eye(s) etc. Comment.
Answer:
Development of organs in an organism is regulated by expression of different sets of genes in a definite sequence in an orderly manner. Any disturbance in co-ordination ‘ and expression of genes will lead to abnormal i. organ formation.
Question 9.
In a nucleus, the number of ribonucleoside triphosphates is 10 times the number of deoxy ribonucleoside triphosphates, but only deoxy ribonucleotides are added during the DNA replication. Suggest a mechanism.
Answer:
Recent studies show that there is a region in the active site cleft of the polymerase enzyme that monitors the 2′ and 3′ substituents of the incoming nucleotide and identifies the sugar component. It ensures that only deoxyribonucleotides are picked up during replication from the nuclear pool.
Question 10.
Name a few enzymes involved in DNA replication other than DNA polymerase and ligase. Name the key functions for each of them.
Answer:
- DNA helicase which stimulates the separation of the two strands.
- Topoisomerase which changes the degree of supercoiling in DNA by cutting one or both strands, which forms
- Primase which forms RNA primer strands on single-stranded DNA templates.
Question 11.
Name any three viruses which have RNA as the genetic material.
Answer:
- Tobacco Mosaic Virus
- Human Immunodeficiency Virus
- Influenza Virus
Short Answer Type Questions
Question 1.
Define transformation in Griffith’s experiment. Discuss how it helps in the identification of DNA as the genetic material.
Answer:
In 1928, Frederick Griffith performed the transformation experiment using Streptococcus pneumoniae. When he injected heat killed, virulent S strain along with non-virulent, live R strain in mice, then the mice died. It showed that something from dead S strain transformed the non-virulent R strain into virulent one. This phenomenon was called transformation by him. Transformation is the phenomenon by which the transforming principle (as named by Griffith), isolated from one type of cell, when introduced into another type, is able to bestow some of the properties of the former to the latter. This discovery started the quest for identification of this transforming principle. Avery, MacLeod and McCarty later purified biochemicals (proteins, DNA, RNA, etc.) from the heat-killed S cells to see which ones could transform live R cells into S cells. They discovered that DNA alone (neither proteins nor RNAs) from S bacteria caused R bacteria to become transformed. They discovered that protein-digesting enzymes (proteases) and RNA-digesting enzymes (RNases) did not affect transformation. Digestion with DNase did inhibit transformation, suggesting that the DNA caused the transformation.
Question 2.
Who revealed biochemical nature of the transforming principle? How was it done?
Answer:
Refer answer 1.
Question 3.
Discuss the significance of heavy isotope of nitrogen in the Meseison and Stahl’s experiment.
Answer:
15N is not a radioactive isotope. It is a heavy isotope of N and can be separated from 14N by density gradient centrifugation. This helped Meseison and Stahl to prove that DNA replicates semiconservatively. Meseison and Stahl extracted bacterial DNA and centrifuged it in caesium chloride solution. Depending on the mass of the molecule, the DNA would settle out at a particular point in the tube. They first grew Escherichia coli bacteria in a medium containing heavy isotope 15N for several generations.
The 15N bacteria were then transferred to a growth medium containing the normal, lighter isotope of nitrogen, 14N, where they reproduced by cell division. Extracts of DNA from the first generation offspring were shown to have a lower density, since half the DNA was made up of the original strand containing 15N and the other half was made up of the new strand containing 14N. At succeeding generation times, the DNA extracts were found to separate at lower densities indicating that have a lower proportion of ,5N as more 14N had incorporated into the bacterial DNA. This was conclusive evidence for the semiconservative method of DNA replication.
Question 4.
Define a cistron. Giving examples, differentiate between monocistronic and polycistronic ‘transcription unit.
Answer:
Cistron is a length of DNA that contains the information for coding a specific polypeptide chain or codes for a functional RNA molecule (i.e., mRNA, transfer RNA or ribosomal RNA).
Monocistronic transcription unit is a type of messenger RNA that can encode only one polypeptide per RNA molecule. In eukaryotic cells, virtually all messenger RNAs are monocistronic.
Polycistronic transcription unit is a type of messenger RNA that can encode more than one pqlypeptide separately within the same RNA molecule. Bacterial messenger RNA is generally polycistronic.
Question 5.
Give any six features of the human genome.
Answer:
Features of the human genome are as follows:
- Human genome has 3.1647 billion nucleotide base pairs.
- The average gene size is 3000 base pairs. The largest gene is that of Duchenne muscular dystrophy on X-chromosome. It has 2.4 million (2400 kilo) base pairs. (3-globin and insulin genes are less than 10 kilobases.
- The human genome consists of about 30,000 genes. Previously, it was estimated to contain 80,000 to 100,000 genes.
- Chromosome 1 has 2968 genes while Y-chromosome has 231 genes. They are the maximum and minimum genes for the human chromosomes.
- The function of over 50% of discovered genes is unknown.
- Less than 2% of the genome represents structural genes that code for proteins.
Question 6.
During DNA replication, why is it that the entire molecule does not open in one go? Explain replication fork. What are the two functions that the monomers (dNTPs) play?
Answer:
Whole of DNA does not open in one stretch due to very high energy requirement. The point of separation proceeds slowly towards both the directions in DNA strands. In each direction, it gives the appearance of Y-shaped structure called replication fork. Deoxyribonucleoside triphosphates (dNTPs) serve dual purposes. In addition to acting as substrates, they provide energy for polymerisation reaction (the two terminal phosphates in a deoxynucleoside triphosphates are high-energy phosphates, same as in case of ATP).
Question 7.
Retroviruses do not follow central dogma. Comment.
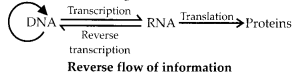
Answer:
Retroviruses follow reverse of central dogma. Temin and Baltimore reported that double stranded RNA of Rous Sarcoma
Virus (RSV) operates a central dogma reverse (inverse flow of information). RNA of these viruses first synthesises DN A through reverse transcription or feminism. DNA then transfers information to RNA which takes part in translation of coded information to form polypeptide. The mechanism is characteristic of retroviruses, e.g., HIV.
Question 8.
In an experiment, DNA is treated with a compound which tends to place itself amongst the stacks of nitrogenous base pairs. As a result of this, the distance between two consecutive bases increases from 0.34 nm to 0.44 nm. Calculate the length of DNA double helix (which has 2 x 109 bp) in the presence of saturating amount of this compound.
Answer:
In the given question, the distance between two consecutive base pairs is 0.44 nm (0.44 x 109m).
Total number of base pair is 2 x 109 bp.
Length of DNA double helix is calculated by multiplying the total number of bp with distance between two consecutive bp i.e., 2 x 109 bp x 0.44 x 10-9 m/bp.Therefore, length of DNA double helix is 0.88 m.
Question 9.
What would happen if histones were to be mutated and made rich in acidic amino acids such as aspartic acid and glutamic acid in place of basic amino acids such as lysine and arginine?
Answer:
Acidic amino acids are negatively charged. DNA is also negatively charged, Therefore, acidic amino acids will not be able to hold DNA over them. This will lead to failure of packaging of DNA material resulting no chromatin formation.
Question 10.
Recall the experiments done by Frederick Griffith, Avery, MacLeod and McCarty, where DNA was speculated to be the genetic material. If RNA, instead of DNA was the genetic material, would the heat killed strain of Pneumococcus have transformed the R-strain into virulent
Answer:
RNA is labile and prone to degradation Recall the experiments done by Frederick Griffith, Avery, MacLeod and McCarty, where DNA was speculated to be the genetic material. If RNA, instead of DNA was the genetic material, would the heat killed strain of Pneumococcus have transformed the R-strain into virulent form.
Question 11.
You are repeating the Hershey-Chase experiment and are provided with two isotopes 32P and 15N (in place of 35S in the original experiment). How do you expect your results to be different?
Answer:
32P is a radioactive isotope but 15N is not. 15N is heavier isotope of nitrogen. Even if it can be detected, 15N will not be able to differentiate between protein and genetic material because it will get incorporated both in DNA as well as proteins. Therefore, the experiment will not give any conclusive result.
Question 12.
There is only one possible sequence of amino acids when deduced from a given nucleotide sequence. But multiple nucleotide sequences can be deduced from a single amino acid sequence. Explain this phenomena.
Answer:
There are 64 triplet codons and only 20 amino acids. Some amino acids are coded by more than one codons. Except tryptophan (UGG) and methionine (AUG) which have single codon each, all other amino acids involved in protein synthesis have more than one codon. In degenerate codons, mostly the first two nitrogen bases are similar while the third one is different. Thus as a codon codes only for a specific amino acid we can deduce a single amino acid sequence from a nucleotide sequence. But, as an amino acid can be coded by multiple codons thus many nucleotide sequences can be coded from a given amino acid sequence.
Question 13.
A single base mutation in a gene may not ‘always’ result in loss or gain of function. Do you think the statement is correct? Defend your answer.
Answer:
Sometimes single base mutation may not cause formation of new amino acid. It occurs usually when change’ occurs in third base of triplet codon, e.g. G G A can be replaced by GGU, GGC, and GGG without affecting the incorporation of amino acid glycine. Such mutations are called silent mutations and do not cause any change in the phenotype of the organism.
Question 14.
A low level of expression of lac operon occurs at all the time. Can you explain the logic behind this phenomenon?
Answer:
The lac operon synthesises the permease enzyme which is responsible for transporting lactose into the cell. If permease will not be present, then the lactose cannot enter the cell and cannot act as inducer for lac operon. Hence, a low level of expression of lac operon is always maintained in the cell, so that whenever lactose is present, it can enter the cell and induce lac operon.
Question 15.
How has the sequencing of human genome opened new windows for treatment of various genetic disorders. Discuss amongst your classmates.
Answer:
Human genome sequencing has been very useful in treatment of genetic disorders, such as:
- It has been found that more than 1200 genes are responsible for common human cardiovascular diseases, endocrine diseases (like diabetes), neurological disorders (like Alzheimer’s disease), cancer and many more. Thus, the medicines that specifically target these genes can be made.
- Efforts are in progress to determine genes that will change cancerous cells into normal.
- All the genes or transcripts in a particular t tissue, organ or tumor can be analysed toknow the cause or effect produced in it.
Question 16.
The total number of genes in humans is far less (< 25,000) than the previous estimate (up to 1,40,000 gene). Comment.
Answer:
At the start of genome sequencing project, scientists estimated that human genome has about 1,40,000 genes. The number gradually fell down to 30,000 and finally has been found to be between 20,000 to 25,000. The estimates were so high because scientists have found E.coli (single celled) to have about 4300 genes and Caenorhabditis elegans (about 1000 cell) to have about 19000 genes. Thus, asper the assumption that the more complex an organism the more is number of genes, the human gene number has been estimated to be more than one lakh. But, number of genes has no relationship with complexity of organism. Besides, it also has been found that genome size and gene number is unrelated because only 2% of the genome actually consists of structural genes, rest 98% is non-coding DNA.
Question 17.
Now, sequencing of total genome is getting less expensive day by the day. Soon it may be affordable for a common man to get his genome sequenced. What in your opinion could be the advantage and disadvantage of this development?
Answer:
Advantages of genome sequencing are as follows:
- It will provide information about genetic abnormalities.
- There will be possibility of transfer of certain desired genes to the progeny.
- Proneness to certain diseases will make a person avoid certain factors causing those diseases.
- Information about timing of normal appearance of degenerative diseases and how to postpone them can be gathered.
- Metobolic defects can be taken care of. Disadvantages of genome sequencing are as follows:
- Knowing about disease much in advance may lead the person under depression.
- People will become keen to improve their genetic constitution.
Question 18.
Would it be appropriate to use DNA probes such as VNTR in DNA fingerprinting of a bacteriophage?
Answer:
Bacteriophages do not have repetitive sequences like VNTR in their genome. Bacterial genome is small in size and with all coding sequences. Therefore DNA fingerprinting cannot be done in bacteriophage.
Question 19.
During in vitro synthesis of DNA, a researcher used 2′, 3′ – dideoxycytidine triphosphate as raw nucleotide in place of 2′ -deoxycytidine. What would be the consequence?
Answer:
2′-3′ dideoxycytidine triphosphate cannot form an ester bond necessary for chainformation of DNA, where as 2′ deoxycytidine phosphate can develop such bond. Chain formation of DNA or polymerisation of DNA will stop due to presence of 2′-3′- dideoxycytidine.
Question 20.
What background information did Watson and Crick have made available for developing a model of DNA? What was their contribution?
Answer:
The information already available with them for developing a model of DNA was –
- Erwin Chargaff’s generalisations about DNA structure e., the purine and pyrimidines are always in equal amounts A + G = T + C, and the amount of adenine is always equal to that of thymine and the amount of guanine is always equal to that of cytosine A = T and G = C.
- X Ray diffraction pictures of crystalline DNA produced by Maurice Wilkins and Rosalind Franklin.
Contributions of Watson and Crick to this background data are:
- DNAis a double helical structure with two chains running in antiparallel direction.
- Complementary base pairing rule
- Semi conservative replication
- Mutations occur due to tautomeric changes in nitrogen bases.
Question 21.
What are the functions of (i) methylated guanosine cap, (ii) poly-A “tail” in a mature RNA?
Answer:
Methylated guanosine cap helps in attachment of mRNA to smaller subunit of ribosome during initiation of translation process.
Poly-A-tail provides longevity to mRNA’s life. Tail length and longevity of mRNA are positively correlated.
Question 22.
Do you think that the alternative splicing of exons may enable a structural gene to code for several isoproteins from one and the same gene? if yes, how? if not, why so?
Answer:
Alternative splicing is a controlled process which causes different mRNAs to be produced from a single gene by including some exons in one mRNA under some conditions, while including other exons under other conditions. This greatly increases the number of proteins that can be encoded by < 2$,000 functional genes of the genome. This alternative splicing of exons is sex-specific, tissue-specific and even developmental stage- specific. By alternative splicing of exons, a single gene may encode for several isoproteins and/or proteins of similar class.
Question 23.
Comment on the utility of variability in number of tandem repeats during DNA fingerprinting.
Answer:
Number and type of tandem repeats are specific for every individual. An individual gets them from two parents, but are unique for the individual. Therefore, they can be used to identify the individuals, and their relatives by comparing the repeats using DNA finger printing process.
Long Answer Type Questions
Question 1.
Give an account of Hershey and Chase experiment. What did it conclusively prove? If both DNA and proteins contained phosphorus and sulphur do you think the result would have been the same?
Answer:
Hershey and Chase experiment is based on the fact that phosphorus is present in DNA but not in the protein, and similarly sulphur is present in proteins but not in DNA. They incorporated radioactive isotope of phosphorus (32P) into phage DNA and that of sulphur (35S) into proteins of a separate phage culture. These phage types were used independently to infect the bacterium Escherichia coli. After some time, this mixture was agitated in a blender to separate the empty phage capsids from the surface of bacterial cells and the two were separated by centrifugation. Hershey and Chase showed that when 32P was used, all radioactivity was associated with bacterial cells and if followed, appeared in the progeny phage.
However, when 35S was used, all radioactive material was limited to phage ‘ghosts’ (empty viral protein coats). These results indicated that the DNA of the bacteriophage and not the protein enters the host, where viral replication takes place. Therefore, DNA is the genetic material of T2 bacteriophage. It directs protein coat synthesis and allows replication to occur. If both DNA and proteins contained phosphorus and sulphur, they could not differentiate or separate protein and DNA and could not have concluded anything.
Question 2.
During course of evolution why DNA was chosen over RNA as genetic material? Give reasons by first discussing the desired criteria in a molecule that can act as genetic material and in the light of biochemical differences between DNA and RNA.
Answer:
A molecule that can act as a genetic material must fulfill the following criteria:
- It should be able to generate its replica (replication).
- It should chemically and structurally be stable.
- It should provide the scope for slow changes (mutation) that are required for evolution.
- It should be able to express itself in the form of ‘Mendelian Characters’.
Though RNA is known to be the genetic material in some viruses and early cells, it is not a very suitable genetic material because 2′ – OH group present in every nucleotide of RNA is a reactive group. It means RNA is highly reactive, labile and easily degradable. RNA functions as an enzyme and is, therefore, reactive and unstable.Uracil present in RNA is less stable as compared to thymine (methyl uracil) of DNA.
Being unstable, RNA mutates at a much faster rate, that is why RNA viruses have shorter life span and mutate and evolve very fast. Such ‘rapid changes are harmful to higher forms of life.
DNA is the genetic material of most of the organisms because
- DNA is chemically less reactive and structurally more stable as its nucleotides are not exposed except when they have to express their effect or to be replicated.
- They are comparatively more stable than RNA. Heat which killed bacteria in Griffith’s experiment did not destroy their DNAs.
- Presence of thymine in DNA instead of uracil, provides stability to DNA.
- Hydrogen bonding between purines and pyrimidines and their stacking make DNA more stable for storage of genetic information.
- DNA is capable of undergoing slow mutations required of genetic material.
- It has a power of repairing.
Since DNA is more stable while RNA is more reactive, both the types of nucleic acids have been retained in genetic expression. DNA which is stable enough not to change with different stages of life cycle, age or with change in metabolism of the organism, is retained as better genetic material for the storage of genetic information. It expresses genetic information by protein synthesis through RNA which is more reactive, exposed for quicker action of protein synthesising machinery and thus is better for the transmission of genetic information.
Question 3.
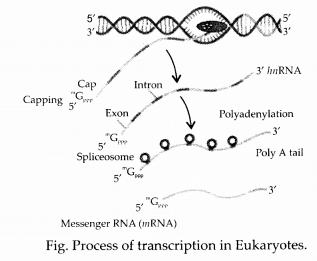
Give an account of post transcriptional modifications of a eukaryotic mRNA.
Answer:
Transcription in eukaryotes occurs within the nucleus. The primary mRNA transcript is longer and localised in the nucleus, where it is also called heterogenous nuclear RNA (/mRNA) or pre-mRNA.
The mRNA is processed from the primary RNA transcript in a process called maturation. Initially, at the 5′ end, a cap (consisting of 7-methyl guanosine or 7 mG) and a tail of poly A at the 3′ end are added. The cap is a chemically modified molecule of guanosine triphosphate (GTP). The primary mRNAs are made up of two types of segments; non¬coding introns and the coding exons. The introns are removed by a process called RNA splicing. Of a pair of small nuclear ribonucleoprotein (snRNPs), one binds to 5′ splice site and the other to 3′ splice site. A spliceosome forms because of interaction between snRNPs and other proteins. This spliceosome uses energy of ATP to cut the RNA and to release the introns. The enzyme ligase joins two adjacent exons to produce mature mRNA.
Question 4.
Discuss the process of translation in detail.
Answer:
The process of decoding of the message from mRNA to protein with the help of fRNA, ribosome and enzyme is called translation (protein synthesis). Protein synthesis occurs over ribosomes. The ribosomes are formed of two subunits. The rosette group formed by ribosomes is called polyribosome. In a polyribosome, ribosomes are held together by strand of /mRNA.
The steps involved in polypeptide synthesis are:
(1) Activation of amino acids : In the presence of ATP, an amino acid combines with its specific aminoacyl-fRNA synthetase.
(2) Charging of fRNA : The complex formed in the above step reacts with tRNA specific for the amino acid to form aminoacyl- tRNA complex.
(3) Initiation : The small subunit of ribosome (with the mRNA) attaches to the large subunit in such a way that the initiation codon (AUG) comes on the P-site of ribosome.
(4) Elongation : Amino acids carried by the tRNA are added one by one at A site of ribosome in the sequence of the codons and become joined together to form
(5) Termination : When the termination codon present on the /mRNA is reached, the polypeptide synthesis stops and the polypeptide is released.
Question 5.
Define an operon. Giving an example, explain an inducible operon.
Answer:
Operon is a functionally integrated genetic unit for the control of gene expression in bacteria, as proposed in the Jacob-Monod hypothesis. Typically, it comprises a closely linked group of structural genes, coding for protein, and adjacent loci controlling their expression-an operator site and a promoter site. Inducible operons are those which normally remain switched off but can be induced to function under certain conditions. Lac operon is an inducible operon. The ; gene of the lac operon codes for a repressor molecule that inhibits synthesis of the structural genes. Lac operon gets switched ‘on’ in the presence of lactose. The repressor molecule coded by i gene is inactivated by interaction with the inducer (lactose). This allows RN Apolymerase access to the promoter, and transcription proceeds. The operon gets switched ‘off’ in the absence of lactose. The repressor molecule binds again with the operator region of the operon and prevents RNA polymerase from transcribing the operon.
Question 6.
There is a paternity dispute for a child’. Which technique can solve the problem. Discuss the principle involved.
Answer:
The paternity dispute of a child can be solved by DNA fingerprinting.
The procedure in DNA-fingerprinting includes the following:
- Extraction – DNA is extracted from the cells in a high-speed, refrigerated centrifuge.
- Amplification – Many copies of the extracted DNA are made by polymerase chain reaction.
- Restriction digestion – DNA is cut into fragments with restriction enzymes into precise reproducible sequences.
- Separation of DNA sequences/restriction fragments – The cut DNA fragments are introduced and passed through electrophoresis containing agarose polymer gel, the separated fragments can be visualised by staining them with a dye that shows fluorescence under ultraviolet radiation.
- Southern blotting – The separated DNA sequences are transferred on a nitrocellulose or nylon membrane.
- Hybridisation – The nylon membrane is immersed in a bath and radioactive probes are added, these probes target a specific nucleotide sequences that is complementary to them.
- Autoradiography – The nylon membrane is pressed on an X-ray film and dark bands develop at the probe sites.
After hybridisation with the radio labelled Variable Number of Tandem Repeats (VNTR) probe and autoradiography, bands of various sizes are formed. The bands form a characteristic pattern which varies from individual to individual. From the patterns developed by the samples A and B it can be confirmed to whether they belong to one individual or two different individuals. If the banding patterns are similar they belong to the same individual but if banding patterns As. are dissimilar then A and B are from different individuals.
Question 7.
Give an account of the methods used in sequencing the human genome.
Answer:
The methods involved two major approaches :
(1) One approach called as Expressed Sequence Tags (ESTs), focussed on identifying all the genes that are expressed as RNA.
(2) Second approach called as sequence annotation, was to simply sequence the whole set of genome, that included all the coding and non-coding sequences and later assigning functions to different regions in the sequences. The steps involved are as follows :
- The total DNA from the cell is isolated and converted into random fragments of relatively smaller sizes.
- These fragments are then cloned in suitable hosts using specialised vectors. The commonly used hosts are bacteria and yeast and the vectors are bacterial artificial chromosomes (BAC) and yeast artificial chromosomes (YAC).
- The fragments are then sequenced using automated DNA sequencers, which work on the principle developed by Frederick Sanger.
- The sequences were then arranged on the basis of certain overlapping regions present in them. This requires the generation of overlapping fragments for sequencing.
- Specialised computer based programmes were developed for alignment of the sequences.
- These sequences were annotated and assigned to the respective chromosomes.
- The next task was to assign the genetic and physical maps on the genome, this was generated using the information on polymorphism of restriction endonuclease recognition sites and certain repetitive DNA, sequences, called microsatellites.
Question 8.
List the various markers that are used in DNA fingerprinting.
Answer:
Various markers used in DNA fingerprinting are as follows:
- Restriction fragment length polymorphism (RFLPs)
- Variable number of tendem repeats (VNTRs)
- Short tendem repeats
- Microsatellite DNA
- Single nucleotide polymorphism (SNPs)
Question 9.
Replication was allowed to take place in the presence of radioactive deoxynucleotide precursors in coli that was a mutant for DNA ligase. Newly synthesised radioactive DNA was purified and strands were separated by denaturation. These were centrifuged using density gradient centrifugation. Which of the following would be a correct result?
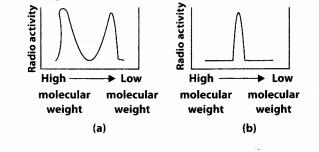
Answer:
(a) Two peaks will be obtained, one of high molecular weight strands that are long, continuous strands which were synthesised on leading strand. The other peak is obtained of low molecular weight strands which is due to short Okazaki fragments synthesised on lagging strand because mutated DNA ligase will not join the Okazaki fragments.
We hope the NCERT Exemplar Solutions for Class 12 Biology chapter 6 Molecular Basis of Inheritance help you. If you have any query regarding .NCERT Exemplar Solutions for Class 12 Biology chapter 6 Molecular Basis of Inheritance, drop a comment below and we will get back to you at the earliest.