By going through these CBSE Class 12 Biology Notes Chapter 11 Biotechnology: Principles and Processes, students can recall all the concepts quickly.
Biotechnology: Principles and Processes Notes Class 12 Biology Chapter 11
→ The European Federation of Biotechnology (FEB) has defined biotechnology as ‘The integration of natural science and organisms, cells, parts thereof, and molecular analogues for products and services. Biotechnology is a science which deals with techniques of using live organisms or enzymes from organisms to produce products and processes useful for man.
All microbes mediated processes, genetically modified organisms, test-tube babies, in vitro fertilization, synthesising a gene, developing a DNA vaccine all processes are a part of biotechnology.
→ Genetic engineering is the science that deals with the synthesis of artificial genes, repair of genes, combining genes from two organisms (recombinant DNA) and manipulation of artificial genes for the improvement of living organisms.
To grow the desired microbe in large quantities for the production of antibiotics, vaccines or enzymes etc., it is very important to maintain microbial-contamination free sterile conditions.
→ For genetically modifying an organism three things are necessary which are:
- modification of DNA,
- the introduction of modified DNA into the host,
- maintenance of introduced DNA in the host and transfer of this DNA in the next progeny.
→ In 1972, Stanley Cohen and Herbert Boyer constructed the first recombinant DNA molecule. They isolated an antibiotic resistance gene and linked it with a native plasmid of Salmonella typhimurium.
→ Plasmids are autonomously replicating circular extrachromosomal DNA. Plasmids are the most widely used cloning vectors. These are double-stranded, circular DNA molecules that can self replicate. A suitable plasmid vector has three properties like low molecular weight, ability to confer readily with selected phenotype traits on host cells and single sites for a large number of restriction endonucleases.
The plasmids carry genes for sexuality, antibiotic resistance etc., but not any vital genes. The cell can survive without them. They can replicate independently of the main genome and being small, can easily come out or get into a cell.
→ Recombinant DNA techniques include DNA at specific sites by using enzymes restriction endonucleases and joining the fragments by enzyme ligases.
It includes the following steps:
- DNA fragments coding for proteins of interest is synthesized chemically or isolated from an organism.
- These DNA fragments are inserted in a restriction endonuclease cleavage site of the vector that does not inactive any gene required for vector’s maintenance.
- The recombinant DNA molecules are now introduced into a host to replicate.
- Recipient host cells that have acquired the recombinant DNA, are selected. Selection pressure is applied to enrich bacteria with a selectable marker.
- Desired clones are then characterised to ensure that they maintain true copies of the DNA segment that was originally cloned.
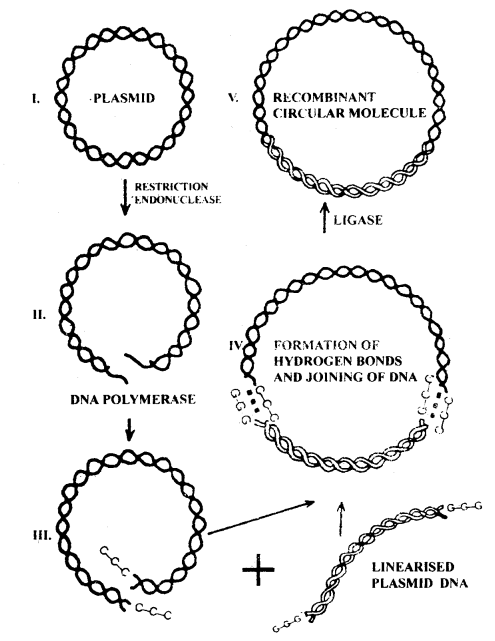
Sequential steps in the formation of recombinant DNA
→ The tools for genetic engineering or recombinant DNA technology are restriction enzymes, polymerase enzymes, biases, vectors and the host organism. After cutting a suitable gene with a restriction enzyme, it is associated with an origin of replication, only then it can multiply itself after insertion into the host genome.
→ Restriction enzymes are also called molecular scissors. In 1968, H.O. Smith, K.W. Wilcoxand T.J. Kelley isolated and characterised the first restriction endonuclease. Hind II from Haemophilus Influenzae bacteria. They found that Hind II always cut DNA at a specific sequence known as recognition sequence. With the help of restriction enzymes, it is possible to cut a DNA sequence. Restriction enzymes belong to class nucleases. These may be of two types-endonucleases and exonucleases.
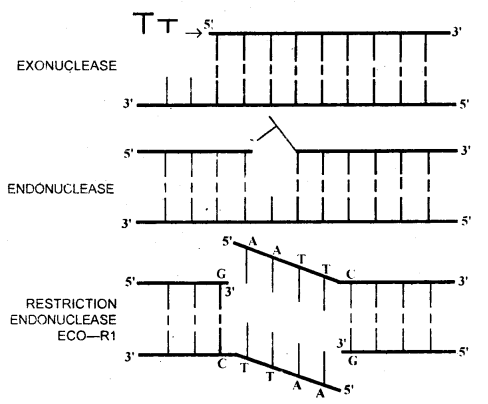
The action of DNA cleaving enzymes
→ Restriction exonucleases, cut the DNA from the ends and restriction endonucleases cut at a specific position within the DNA, e.g. Restriction endonuclease ECORI will cut DNA only if sequence:
5′ — GAATTC — 3′
3′ — CTTAAG — 5′
is present. It cuts the DNA between bases G and A only when sequence GAATTC is present in the DNA (shown in the figure above)
→ The restriction enzyme will act on both the strands and produce a break. If the vector and the source DNA are cleaved using the same restriction enzyme they will have the same kind of sticky ends which can be joined by DNA ligases to produce a recombinant vector.
→ When restriction enzymes make a cut in the DNA strand, it will leave single-stranded portions at the ends. These are overhanging, called sticky ends. Sticky ends can form hydrogen bonds with their complementary cut counterparts by DNA ligases.
→ Palindromes are DNA sequences with base pairs that read the same on the two strands when the orientation of reading is kept the same. For example:
5′ — GAATTC — 3′
3′ — CTTAAG — 5′
In both the strands if read from 5? direction it will be the same. These are the sites for restriction enzymes.
→ After cutting the DNA with restriction enzymes, the fragments of DNA are separated by gel electrophoresis. DNA molecules being negatively charged move towards an anode under an electric field. The matrix used is a natural polymer, agarose. It acts as a sieve and DNA fragments separate according to their size.
DNA fragments can be seen after staining with ethidium bromide, an orange coloured bands when exposed to UV light. The separate strands can be cut from agarose gel and eluted by using various techniques. The selected DNA fragments, purified by gel electrophoresis are used for recombinant DNA construction.
→ Cloning vectors: The vector is an agent which is used to transfer DNA into the host cell. For example plasmid, bacteriophages. The vector is cut with the same restriction endonuclease which was used for chromosomal DNA fragments. The linearized vector and chromosomal DNA fragments are joined together by DNA ligases. Those plasmids which contain an inserted DNA fragment are called recombinant plasmids.
→ Similar to plasmids, bacteriophages can replicate within the bacterial cell independently of the control of chromosomal DNA.
Sometimes there are several copies of plasmids or a high number of bacteriophages per cell if an airy foreign DNA segment is inserted into these vectors a large number of a selected gene can be produced.
To attach a desirable gene to these vectors, the vectors are to be modified: The inserted gene must be attached with an origin of replication to start replication in the host cell, the vector needs a selectable marker so as to identify the recombinant vector. For example, a normal E.Coli cell does not carry resistance to antibiotics like tetracycline, ampicillin etc. so if the cells show resistance towards these antibiotics they are suitable recombinant vectors with suitable markers.
→ To link the desirable DNA with the vector, one or very few recognition sites for restriction enzymes is required. One recognition site will linearise the vector, several recognition sites will complicate the process. The recombinant vectors from non-recombinant vectors can be selected by a replica plating process or by using enzyme β-galactosidase activity on its colour producing substrate. The non-recombinants will produce colour due to active β-galactosidase and recombinants do not produce any colour.
→ For the purpose of clearing genes in plants and animals some vectors like disarmed (cancerous) retroviruses and modified (tumour inducing) plasmid of Agrobacterium tumefacient are used to deliver desirable genes into animals and plants cells respectively.
→ DNA is a hydrophilic molecule that cannot pass through cell membranes. The bacterial cells are made competent or forced to take up DNA. It can be done by the specific concentration of divalent ions like calcium (Ca2+) which increases the efficiency of DNA uptake through cell wall pores. Then the cells are incubated on ice followed by heat shock (at 42°C) and then back to ice. Other ways are microinjection i.e.; direct injection of recombinant DNA by animal cell by using a small needle. Gene gun or biolistics is used for plant cells, cells are bombarded with high-velocity micro gold or tungsten particles coated with DNA.
→ Polymerase chain reaction (PCR) is used to produce many copies of the same gene without using cells. The DNA is incubated in a test tube with DNA polymerases and a mixture of deoxy-ribonucleotides and primers. PCR can make billions of copies of DNA in few hours whereas using bacteria for the same thing will take days. It is the best way to detect an elusive infection, rapid prenatal diagnosis of genetic disorders, and even identification of criminals from tiny samples of blood, tissue or semen by amplification and identification.
→ Bioreactors provide the optimum conditions for getting the desired product by providing optimum growth conditions like temperature, pH, substrate, salts, vitamins and oxygen etc. Bioreactors can be aerobic, submerged, surface and anaerobic type. The most commonly used is the stirring type. A stirrer type bioreactor has an agitator system, an oxygen delivery system, a foam control system, a temperature control system, a pH control system, sampling pouts to withdraw small volumes of culture.
→ Genetic engineering: It is a process by which manipulation of the genetic material of an organism is done.
→ Biotechnology: It is the use of living organisms or of substances obtained from them in the industrial process.
→ Bioreactor: The fermentation tank where fermentation is carried out in the presence of micro-organisms.
→ Broad-spectrum antibiotics: These are the antibiotics that have the ability to act on several pathogenic species.
→ Hybridomas: The hybrid cells of the same clone themselves are known as hybridomas.
→ Monoclonal antibody: The same type of antibody produced by the same clone of all the hybrids.
→ Restriction enzyme: An endonuclease that recogniseS Specific nucleotide sequences in DNA and then makes a double-strand cleavage of DNA molecule.