Here we are providing Class 12 Biology Important Extra Questions and Answers Chapter 6 Molecular Basis of Inheritance. Important Questions for Class 12 Biology are the best resource for students which helps in Class 12 board exams.
Class 12 Biology Chapter 6 Important Extra Questions Molecular Basis of Inheritance
Molecular Basis of Inheritance Important Extra Questions Very Short Answer Type
Question 1.
Name the genetic material for the majority of organisms.
Answer:
DNA (Deoxyribose nucleic acid)
Question 2.
List the function of RNA.
Answer:
RNA acts as genetic material in viruses and also functions as an adapter, structural, and in some cases as a catalytic molecule.
Question 3.
How many nucleotides are present in a bacteriophage Φ × 174?
Answer:
5386.
Question 4.
List the number of base pairs in:
(i) lambda bacteriophage
Answer:
48502 bp
(ii) E.coli and
Answer:
4.6 × 106bp
(iii) haploid content of human DNA.
Answer:
3.3 × 109bp.
Question 5.
Comment two chains of DNA have antiparallel polarity.
Answer:
If one chain has 5′ → 3′ polarity, the other chain has 3′ → 5′ polarity.
Question 6.
What is the difference between DNA and DNAase?
Answer:
DNAs are the number of molecules of DNA and DNAase is an enzyme that digests DNA.
Question 7.
What made DNA as genetic material?
Answer:
As RNA was unstable, it evolved further with certain chemical modifications and formed DNA. DNA became more stable.
Question 8.
What is the average rate of polymerization?
Answer:
2000 bp per second.
Question 9.
List three components of the transcription unit.
Answer:
- A promoter,
- The structural gene,
- A terminator.
Question 10.
What is the term used for fully processed hn RNA?
Answer:
A messenger RNA (mRNA).
Question 11.
What is splicing?
Answer:
It is the removal of introns and the joining of exons in a definite manner.
Question 12.
Where are UTRs present in mRNA strand?
Answer:
UTRs (Untranslated region) are present at both 5′ end (before start codon) and at the 3′ end (after stop signal).
Question 13.
Write the significance of UTRs.
Answer:
They are required for the regulation of an efficient translation process.
Question 14.
Name any two non-sense codons (stop signal).
Answer:
- UGA (Opal)
- UAA (Ochre).
Question 15.
What are the exceptions to the general rule that DNA is the genetic material in all organisms? Give evidence that supports these exceptions.
Answer:
Some animal viruses and all plant viruses contain RNA as their genetic material.
Question 16.
What is a replication fork? (CBSE, Delhi 2011)
Answer:
When replication starts the two strands of DNA unwind to form a Y-shaped structure called the replication fork.
Question 17.
Name the technique by which Gene expression can be controlled with the help of RNA molecule. (CBSE Sample Paper 2018-19)
Answer:
Northern blotting
Question 18.
Name the parts ‘A’ and ‘B’ of the transcription unit given below. (C.B.S.E. Delhi 2008)
Answer:
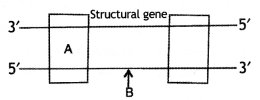
A-Promoter
B-Coding strand.
Question 19.
Mention the role of the codons AUG and UGA during protein synthesis. (CBSE 2011, 2016)
Or
Mention two functions of codon AUG. (CBSE 2010)
Answer:
- AUG acts as a start signal and also code for methionine amino acid.
- UGA acts as a stop signal.
Question 20.
How do histones acquire positive charge? (CBSE Delhi 2011)
Answer:
A histone protein acquires a positive charge because they are rich in basic amino acid residues such as lysine, arginine, and histidine. All are positively charged in their side chains.
Question 21.
Why do DNA fragments move towards the anode during gel electrophoresis? (CBSE Delhi 2018C)
Answer:
DNA molecule is negatively charged. When placed in an electric field it moves towards a positively charged anode.
Question 22.
Name one amino acid which is coded by only one codon. (CBSE Delhi 2018 C)
Answer:
Methionine – AUG/ Tryptophan – UGG
Molecular Basis of Inheritance Important Extra Questions Short Answer Type
Question 1.
If the sequence of coding strand In a transcription unit Is written as follows: (CBSE Delhi 2011)
5′- A T G C A T G C A T G C A T G C A T G C A T G C A T G C – 3′
Write down the sequence of mRNA.
Answer:
The sequence of mRNA shall be:
5′ – U A C G U A C G U A C G U A C G U A C G U A C G U A C G – 3′.
Question 2.
What is Chargaff rule? (CBSE Delhi 2011)
Answer:
Chargaff Rules:
- In DNA molecule, A — T base pairs equals in number to G — C base pairs.
- A + G = T + C, i.e. purines and pyrimidines equal in amount.
- A = T and C = G (Amount).
- The base ratio A + T/G + C may vary from one species to another but is constant for each species. It helps in identifying the source of DNA.
- The deoxyribose sugar and phosphate component occur in equal proportions.
Question 3.
(a) Name the component of a nucleotide responsible for giving 5’— 3′ polarity to a polynucleotide.
Answer:
A polymer has at one end a free phosphate moiety at 5′- end of ribose sugar which is referred to as 5′-end of a polynucleotide chain. Similarly, at the other end of the polymer, the ribose has a free 3-OH group which is referred to as the 3′-end of a polynucleotide chain.
(b) Where in a nucleotide is the glycosidic bond present? (CBSE Delhi 2019 C)
Answer:
A nitrogenous base is linked to pentose sugar through an N-glycosidic linkage to form nucleoside.
Question 4.
Which property of DNA double helix led Watson and Crick to hypothesize a semi-conservative model of DNA replication? Explain.
Or
Write a note on the semi-conservative mode of DNA replication. (CBSE Delhi 2008; Delhi 2011, 2014)
Answer:
The semi-conservative model of DNA replication hypothesized by Watson and Crick was based upon the property that during replication, the two strands would separate and act as a template for the synthesis of new complementary strands. After the completion of replication, each DNA molecule would have one parental and one newly synthesized strand. This scheme was termed as ‘Semi-conservative’ DNA replication.
Question 5.
RNA was the first genetic material, DNA evolved later on. Explain.
Or
Why is RNA considered the first genetic material? (CBSE, 2009 2012)
Answer:
RNA was the first genetic material. There is now enough evidence to suggest that essential life processes (such as metabolism, translation, splicing, etc.) evolved around RNA. RNA used to act as a genetic material as well as a catalyst (there are some important biochemical reactions in living systems that are catalyzed by RNA catalysts and not by protein enzymes). But, RNA being a catalyst was reactive and hence unstable. Therefore, DNA has evolved from RNA with chemical modifications that make it more stable. DNA being double-stranded and having complementary strands further resists changes by evolving a process of repair.
Question 6.
Discuss the significance of the heavy isotope of nitrogen in Meselson and Stahl’s experiment.
Answer:
- By using the heavy isotope of nitrogen, they could find out the semiconservative nature of DNA replication as the densities of DNA having 15N in both the strands 15N/14N DNA and 14N/14N DNA were all different.
- The hybrid DNA (15N/14N DNA) had a density intermediate between that of heavy DNA (15N/15N DNA) and that of light/normal DNA (14N/14N DNA).
Question 7.
Differentiate polycistronic mRNA and monocistronic mRNA.
Answer:
Differences between polycistronic mRNA and monocistronic mRNA:
| Polycistronic mRNA | Monocistronic mRNA |
| 1. It is the mRNA that can code for only one polypeptide, i.e. it has one cistron | 1. It is the mRNA that can code for more than one polypeptide, i.e. it has more than one cistron. |
| 2. It is normally found in eukaryotic cells. | 2. It is found in prokaryotic cells. |
Question 8.
You are repeating the Hershey-Chase experiment and are provided with two isotopes 32p and 15N (in place of 35S in the original experiment). How do you expect your results to be different?
Answer:
- The use of 15N will not give any conclusive result because it is only a heavy isotope of nitrogen.
- In the original experiment, 35S was detected only in the supernatant as it was incorporated in protein only, while 15N will be incorporated into proteins as well as in DNA and hence it would appear both in the supernatant and in the sediment as well.
Question 9.
What is DNA fingerprinting? Mention its applications.
Or
List any two applications of the DNA fingerprinting technique. (CBSE Delhi 2018)
Answer:
DNA fingerprinting: The technique of using DNA fragments, resulting from restriction endonuclease enzyme cleavage to identify particular individuals, is called DNA fingerprinting.
Applications of DNA Fingerprinting:
- Paternity disputes can be solved by DNA fingerprinting.
- It can solve the problems of evolution.
- It can be used to study the breeding patterns of animals facing the danger of extinction.
- It is useful in restoring the health of the patients suffering from leukemia (blood cancer).
- It is very useful in the detection of crime and legal pursuits.
Question 10.
What are the major enzymes of DNA replication? (CBSE 2009)
Answer:
Enzymes of DNA Replication:
- DNA-dependent DNA polymerase. It catalyzes the polymerization of deoxynucleotides,
- Okazaki fragments, which are then quickly joined together by an enzyme known as DNA ligase.
- The discovery of helicases and topoisomerase enzymes explains the unwinding of the DNA helix.
Question 11.
One of the codons on mRNA is AUG. Draw the structure of the tRNA adapter molecule for this codon. Explain the uniqueness of this tRNA. (CBSE Delhi and Outside Delhi, 2008)
Answer:
Transfer RNA or soluble RNA or Adapter RNA (tRNA or sRNA). It constitutes 15% of total RNA and is the smallest out of three with only 70-85 nucleotides having a sedimentation coefficient of 45.
It is unique because it has double specificity, one for codon on mRNA strand and the other for the corresponding amino acid.
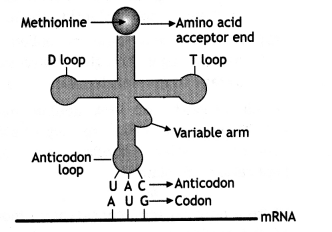
Structure of tRNA adapter
Question 12.
Briefly describe the termination of a polypeptide chain. (C8SE 2009)
Answer:
Termination of polypeptide synthesis:
- When one of the termination codons (UAA, UAG, UGA) comes at the A-site, it does not code for any amino acid and there is no tRNA molecule for it.
- As a result, the polypeptide synthesis (or elongation of the polypeptide) stops.
- The polypeptide synthesized is released from the ribosome, catalyzed by a ‘release factor’.
Question 13.
Write the dual purpose served by Deoxyribonucleoside triphosphates in polymerization. (CBSE Delhi 2018)
Answer:
Deoxyribonucleoside triphosphates serve as substrates, i.e. nucleotides during replication, and also provide energy for polymerization reaction by cleavage of high energy terminal phosphates bond. Thus they serve dual purposes.
Question 14.
Although a prokaryotic cell has no defined nucleus, yet DNA is not scattered throughout the cell. Explain. (CBSE Delhi 2018)
Answer:
In the nucleoid region, DNA that is negatively charged is held with some proteins that are positively charged. The DNA in the nucleoid is organized in large loops held by proteins. And the DNA in the form of single chromosomes is attached to the mesosome at a point. 15
Question 15.
Differentiate between the genetic codes given below:
(i) Unambiguous and Universal
Answer:
- Unambiguous: The code is specific, i.e. one Condon code for onLy one amino acid.
- Universal: The code is the same in aLL organisms.
(ii) Degenerate and Initiator (CBSE Delhi 2017)
Answer:
- Degenerate: When an amino acid is coded by more than one codon, it is said to be degenerate.
- Initiator: AUG is an initiator codon, i.e. it initiates the translation process and also codes for methionine.
Question 16.
Why does the lac operon shut down sometime after the addition of lactose in the medium where E.coti was growing? Why low-level expression of the lac operon is always required? (CBSE Sample Paper 2018-19)
Answer:
After the addition of lactose, a complete breakdown of lactose to glucose and galactose takes place. Therefore, there is no more lactose to bind to the repressor protein and the lac operon shuts down.
A very low level of expression of lac operon has to be present in the cell all the time, otherwise, lactose cannot enter the cells.
Question 17.
Carefully examine structures A and B of pentose sugar given below. Which one of the two is more reactive? Give reasons. (CBSE Sample Paper 2019-20)
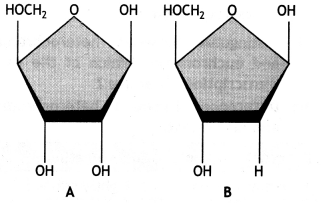
Answer:
The pentose sugar of figure A is more reactive.
Reasons:
- There are two -OH groups present in the pentose sugar.
- It makes it more labile.
- It is easily degradable.
Molecular Basis of Inheritance Important Extra Questions Long Answer Type
Question 1.
(i) How are the following formed and involved in DNA packaging in a nucleus of a cell?
(a) Histone octamer
(b) Nucleosome
(c) Chromatin
Answer:
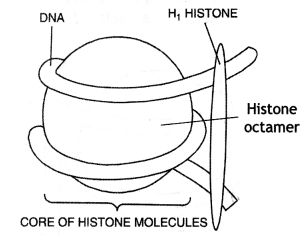
Packaging of DNA.
(a) Histone octamer: Five types of histone proteins (H1, H2A, H2B, H3, and H4) are involved. Out of these, four of them H1 A, H2 B, H3, and H4 occur in pairs to produce histone octamer also called the nu body. Histones are organized in a form of a compact unit formed of 8 molecules hence called histone octamer.
(b) Nucleosome: The unit of compaction of DNA is the nucleosome. About 146 bp of DMA is wrapped
over histone octamer for 1 \(\frac{3}{4}\) turn to form nucleosome of the size 110 × 60 A.
(c) Chromatin: Linker DNA connects two adjacent nucleosomes. It bears H1 protein. As a result, a chain is formed called chromatic. Nucleosome chain gives ‘beads on string’ appearance under the electron microscope. Chromatin are repeating units of a structure located on the nucleosome.
(ii) Differentiate between Euchromatin: 1 and Heterochromatin. (CBSE Delhi 2016)
Answer:
| Heterochromatin | Euchromatin |
| 1. Darkly stained. | 1. lightly stained. |
| 2. Condensed regions of chromatin fibers. | 2. less tightly coiled regions of chromatin fibers. |
| 3. Transcriptionatty inactive or less active. | 3. Transcnptionally active. |
| 4. less affected by temperature, sex, or age. it is not acetylated. | 4. More affected by temperature, sex, or age. It is acetylated during interphase. |
Question 2.
Distinguish between heterochromatin and euchromatin. Which of the two is transcriptionally active?
Answer:
Differences between heterochromatin and euchromatin:
| Heterochromatin | Euchromatin |
| 1. Darkly stained. | 1. lightly stained. |
| 2. Condensed regions of chromatin fibers. | 2. less tightly coiled regions of chromatin fibers. |
| 3. Transcriptionatty inactive or less active. | 3. Transcnptionally active. |
| 4. less affected by temperature, sex, or age. it is not acetylated. | 4. More affected by temperature, sex, or age. It is acetylated during interphase. |
Euchromatin is more active transcriptionally.
Question 3.
Write a note on messenger RNA.
Answer:
Messenger RNA (mRNA).
It forms only 5% of total RNA but is the longest of all. It brings instructions from DNA for the formation of a particular polypeptide. The instructions are coded in the form of a base sequence called genetic code. Three adjacent nitrogen bases specify a particular amino acid. The formation of polypeptides occurs over the ribosomes. mRNA gets attached to ribosomes.
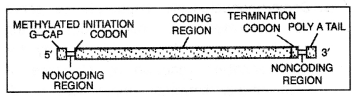
mRNA.
It starts as a cap for attachment with the ribosome. It is followed by an initiation codon (AUG) either immediately or after a small non-coding region. It is followed by the coding region followed by the termination codon (UAA, UAG, and UGA). Then there is a small non-coding region and poly-A area at 3 termini. The mRNA may specify only a single polypeptide or a number of them called monocistronic and polycistronic respectively.
The life span of mRNA maybe a few minutes to an hour or even days in the case of RBC.
Question 4.
What is genetic code? List the properties of genetic code.
Answer:
The code language of DNA and mRNA is complementary. So, genetic code is the sequence of nucleotides in DNA and RNA that determines the amino acid sequence in proteins. Some amino acids are specified by more than one codon. The sequence of nucleotide on the tRNA molecule which complements the codon is called anticodon, e.g. one of the codons for the amino acid leucine is CUG and the anticodon is GAC. Similarly, the codon for phenylalanine is UUU, while the anticodon is AAA.
Properties of genetic code:
The following properties of genetic code have now been proved by experimental evidence.
- The code is a triplet.
- The code is degenerate.
- The code is non¬overlapping.
- The code is commaless.
- The code is non-ambiguous.
- The code is universal,
- Collinearity.
Both polypeptide and DNA or mRNA have a linear arrangement of their components.
The term code letter stands for a nucleotide A, T, G, or C in DNA and A, U, G, or C in RNA. The sequence of three does not code for any amino acid, such codons are called a non-sense codon, e.g. UGA.
Question 5.
What is the role of ribosomes during translation? Ribosomes move along mRNA molecules and catalyze the assembly of amino acids into protein chambers. (CBSE Outside Delhi 2019)
Answer:
Role of ribosomes: Ribosomes usually form linear or helical groups during active protein synthesis called polyribosomes or polysomes. The mRNA strand having coded information joins along with smaller subunits of ribosomes. The adjacent ribosomes are 360 A apart.
The different parts of the ribosome connected with protein synthesis are:
- A tunnel for mRNA.
- A groove for the passage of newly synthesized polypeptide (larger subunit).
- Two active sites (P-site-peptidyl transfer or donor site and A-site or aminoacyl or acceptor site).
- A binding site for tRNA near A-site.
- Presence of enzyme peptidyl transferase.
- Recognition point of smaller subunit for mRNA.
- Presence of GTP-ase, binding sites for elongation factors, and translocases.
Question 6.
Describe the steps in the sequencing of the human genome.
Answer:
Sequencing of a genome.
The method involved two major approaches:
- Expressed Sequence Tags (ESTs): It is focused on identifying all the genes that are expressed as RNAs.
- Sequence Annotation: It involves simply sequence the whole set of the genome that included all the coding and non-coding sequences and then assigning functions to different regions in the sequence.
HGP followed the second technique:
- The total DNA from the cell is isolated and converted into random fragments of relatively smaller sizes.
- These fragments are then cloned in suitable hosts using specialized vectors; the commonly used hosts are bacteria and yeast and the vectors are bacterial artificial chromosomes (BAC) and yeast artificial chromosomes (YAC).
- The fragments are then sequenced using automated DNA sequences.
- The sequences were then arranged on the basis of certain overlapping regions present in them; this required the generation of overlapping fragments for sequencing.
- These sequences are annotated and assigned to the respective chromosomes.
Question 7.
(i) Why is DNA molecule a more stable genetic material than RNA? Explain.
Answer:
There is now enough evidence to suggest that essential life processes (such as metabolism, translation, splicing, etc.) revolved around RNA. RNA used to act as a genetic material as well as a catalyst (there are some important biochemical reactions in living systems that are catalyzed by RNA catalyst and not by protein enzymes). But, RNA being a catalyst was reactive and hence unstable. Therefore, DNA has evolved from RNA with chemical modifications that make it more stable. DNA being double-stranded and having complementary strands further resists changes by evolving a process of repair.
(ii) ‘Unambiguous’, ‘degenerate’ and ‘universal’ are some of the salient features of genetic code. Explain. (CBSE 2008, 2011, Delhi 2014, 2016)
Answer:
Unambiguous: Each codon codes for one amino acid, none for more than one.
Degenerate: One amino acid often has more than one triplet codon; only methionine and tryptophan have a single triplet codon.
Universal: The genetic code is universal. A given codon in DNA and m RNA specifies the same amino acid in all organisms from viruses, bacteria to human beings.
Question 8.
Explain the role of tRNA in the initiation of protein synthesis. (CBSE 2012)
Answer:
Role of tRNA’ in the initiation of protein synthesis:
- tRNA is an adapter molecule that on one hand would bind to a specific amino acid.
- The tRNA then called sRNA (soluble RNA).
- tRNA has an anticodon loop that has bases complementary to the code.
- It has an amino acid acceptor end to which it binds to an amino acid.
- tRNAs are specific for each amino acid.
- For initiation, there is another tRNA, that is referred to as initiator tRNA.
- Amino acids are activated in the presence of ATP and joined to their corresponding tRNA it is called charging of tRNA or aminoacylation of tRNA.
- Two such charged tRNAs are brought close and the formation of peptide bond occurs.
Question 9.
List the criteria that can act as genetic material must fulfill. Which one of the criteria is best fulfilled by DNA or RNA thus making one of them a better genetic material? Explain. (CBSE (Delhi) 2016)
Answer:
Essential requirements of genetic material:
- A genetic material should be able to store and express information to confer the heritable characters of living organisms.
- It should be capable to make its own replica.
- Genetic material should also have the mechanism to undergo mutations that will generate variations.
Question 10.
A small stretch of DNA strand that codes for a polypeptide are shown below:
3’— — — — CAT CAT AGA TGA AAC — — — — 5’
(a) Which type of mutation could have occurred in each type resulting in the following mistakes during rep¬lication of the above original se¬quence?
(i) 3’ … … … … CAT CAT AGA TGA ATC … … … 5’
Answer:
A point mutation (single base substitution)
(ii) 3’ … … … … CAT ATA GAT GAA AC … … … 5’
Answer:
A point mutation (single base deletion)
(b) How many amino acids will be translated from each of the above strands (i) and (ii)? (CBSE Sample Paper 2019-20)
Answer:
(i) 4 amino acids
(ii) 4 amino acids
Question 11.
(i) In the human genome which one of the chromosomes has the most genes and which one has the fewest?
Answer:
Chromosome 1 has the most genes (2968) and Y-chromosome has the least gene (231).
(ii) Scientists have identified about 1.4 million single nucleotide polymorphs in the human genome. How is the information of their existence going to help the scientists? (CBSE2009)
Answer:
This information promises to revolutionize the processes of finding the chromosomal location for disease-associated sequences and tracing human history.
Question 12.
Describe the structure of an RNA polynucleotide chain having four different types of nucleotides. (CBSE Delhi 2013, 2019)
Answer:
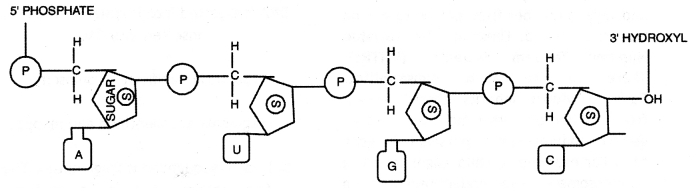
RNA polynucleotide
RNA polynucleotide:
- RNA is formed of polynucleotides of ribose series.
- Each nucleotide is formed of a nitrogen base, ribose sugar (pentose), and phosphoric acid.
- There are two types of nitrogen bases-purines and pyrimidines.
- Adenine (A) and Guanine (G) are purines.
- Cytosine (C) and Uracil (U) are pyrimidines.
- The nitrogen base is linked to pentose sugar by N-glycosidic linkage to form nucleoside.
- When a phosphate group is linked to 5′ -OH of a nucleoside through phosphodiester linkage thus forms nucleotide.
- Nucleotides of ribose series present are AMP, GMP, CMP, and UMP.
- Two nucleotides are linked through 3′ – 5′ phosphodiester linkage to form dinucleotides.
- More nucleotides can be joined in such a manner to form a polynucleotide chain.
Question 13.
Describe the initiation process of transcription in bacteria. (CBSE 2010, 2016, 2019)
Answer:
Initiation of transcription in bacteria:
- The process of copying genetic information from antisense or template strands of DNA into RNA is called transcription.
- The segment of DNA that takes part in transcription is called the transcription unit. It has three components
(a) a promoter,
(b) the structural gene and
(c) a terminator. - The structural gene is composed of that strand of DNA that has 3′ → 5′ polarity as transcription can occur only in the 5′ → 3′ direction.
- Transcription requires a DNA-dependent-RNA polymerase and initiation factor.
- Bacteria have only one type of RNA polymerase which transcribes all three types of RNAs.
- Ribonucleotides of ribose series are activated through phosphorylation (ATP, GTP, CTP, UTP).
- Transcription begins at the initiation site. A promotor has an RNA polymerase recognition site and it binds to the specific site.
- Enzymes required for the unwinding of the chain are unwound and single-stranded binding proteins.
- Nucleotides are added as per the base-pairing rule.
Question 14.
State the role of VNTRs in DNA fingerprinting. (CBSE Outside Delhi 2013)
Answer:
Role of Variable Number of Tandem Repeats (VNTRs) Short nucleotide repeats in the DNA are very specific in each individual and vary in number from person to person but are inherited. These are the ‘Variable Number Tandem Repeats’ (VNTRs). These are also called ‘minisatellites. Each individual inherits these repeats from his/her parents which are used as genetic markers in a personal identity test.
For example, a child might inherit a chromosome with six tandem repeats from the mother and the same tandem repeated four times in the homologous chromosome inherited from the father. The half of VNTR alleles of the child resemble that of the mother and half that of the father.
Question 15.
(i) List the two methodologies which were involved in the human genome project. Mention how they were used.
Answer:
(a) Expressed Sequence Tags (ESTs): This method focuses on identifying all the genes that are expressed as RNA.
(b) Sequence Annotation: It is a method of simply sequencing the whole set of the genome that contains all the coding and non-coding sequences, and then assigning different regions in the sequence with functions.
(ii) Expand ‘YAC’ and mention what it was used for. (CBSE Delhi 2017)
Answer:
‘YAC’ is an abbreviated form of ‘Yeast Artificial Chromosome’. It is used as a cloning vector for cloning DNA fragments in a suitable host so that DNA sequencing can be done.
Question 16.
Summarise the process by which the sequence of DNA bases in the Human Genome Project was determined using the method developed by Frederick Sanger. Name a free-living non-pathogenic nematode whose DNA has been completely sequenced. (CBSE Sample Paper 2019-20)
Answer:
(a) The HGP strategy:

(b) Chromosome 1
(c) Caenorhabditis elegans
Question 17.
Why is a DNA molecule considered a better hereditary material than an RNA molecule? (CBSE Delhi 2018C
Answer:
DNA is the better hereditary material compared to RNA because of the following features:
- DNA molecule is more stable tha> RNA (as thymine is present here instead of uracil found in RNA).
- DNA is less reactive than RNA (a- does not have 2’ OH group wl makes RNA very reactive).
- Since DNA is less reactive so it is t easily degradable.
- Chances of mutation are less. compared to RNA.
Question 18.
Name the three RNA polymerases found in eukaryotic cells and mention their functions. (CBSE Delhi 2018C)
Answer:
- RNA polymerase I – It transcribes ribosomal RNAs (28S, 18S, 5.8S rRNAs)
- RNA polymerase II – It transcribes precursors of mRNA – heterogeneous nuclear RNA (hnRNA)
- RNA polymerase III – It transcribes transfer RNA (tRNA), 5SrRNAand small nuclear RNAs (snRNAs)
Question 19.
Explain the post-transcriptional modifications the hn-RNA undergoes in eukaryotic cells. (CBSE Delhi 2018C)
Answer:
In eukaryotes, the primary transcript is not functional due to the presence of exons and introns. Therefore it requires certain modifications to make it functional. First of all, introns are removed and the remaining exons are joined together in order by the process called Splicing.
Then the hnRNA undergoes capping followed by tailing. In capping, methyl guanosine triphosphate is added to the 5’ end of hnRNA. In tailing 200 -300 adenylate residues are added to 3’ end in a template-independent manner. Now the hnRNA has been processed into mRNA.
Question 20.
What are the aims of bioinformatics?
Answer:
Aims of bioinformatics:
- To spread scientifically investigated knowledge for the benefit of the research community.
- To transform the biological polymeric sequences into sequences of digital symbols and to store them as databases.
- To develop a variety of methods and tools of software for data analysis.
Question 21.
Describe Griffith’s experiment to demonstrate that DNA is the basic genetic material. What explanation for Griffith’s observation was given by Avery, McCarty, and MacCleod? (CBSE Delhi 2008, 2009, 2012)
Answer:
Griffith’s experiment demonstrates DNA as genetic material. Transformation experiments were initially conducted by F. Griffith in 1928.
- He injected a mixture of two strains of Pneumococcus (Diplococcus pneumoniae) into mice. One of these two strains S III was virulent and the other strain All was non-virulent.
- The S III type bacteria when injected into mice cause pneumonia and ultimately to death. The R II type bacteria when injected, no pneumonia occurred.
- The S III bacteria, prior to injection are killed by heating. It is then injected into the mice. It did not cause the disease.
- Heat killed S III and RII (non-virulent) bacteria, when injected into mice, causing pneumonia. Finally, death occurred.
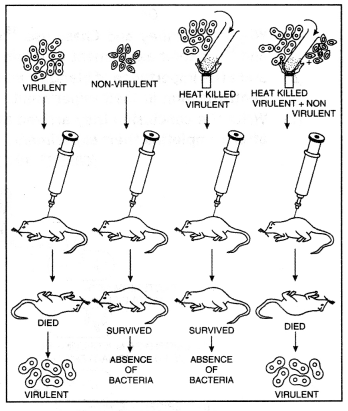
Griffith’s experiment demonstration transformation in Pneumococcus.
This proved that the DNA of S III type bacteria has transformed the DNA of R II type bacteria into virulent type S III. This phenomenon of transferring characters of one strain to another by using a DNA extract of the former is called transformation. Conclusion. Griffith concluded that virulence was transferred from S-Type dead cells to R-Type living cells in the form of a capsule (some component of a cell) rather than the whole cells.
Contribution of Avery, MacCleod, and MacCarty: Avery, MacCleod, and McCarty gave the proof that the “transforming agent” is DNA. They carried out the experiments with Diplococcus and showed the transformation of type R-ll to type S-III. They found that proteases and RNases did not affect transformation but DNAses affect it. Thus they gave proof that the active component was DNA and not the RNA, proteins, or polysaccharides.
Question 22.
How did Hershey and Chase differentiate between DNA and protein in their experiment while proving that DNA is the genetic material? (CBSE 20015)
Or
Why did Hershey and Chase use 35S and 32P in their experiment? Explain. State the importance of blending and centrifugation in their experiment. Write the conclusion they arrived at after completing their experiment. (CBSE 20019 C)
Or
Hershey and Chase carried out their experiment under three steps: (a) Infection, (b) Blending, and (c) Centrifugation.
Or
Explain each one of these steps that helped them to prove that DNA is the hereditary material. (CBSE (Outside Delhi) 2019)
Answer:
1. Alfred Hershey and Martha Chase (1952) worked with viruses that infect bacteria called bacteriophages.
2. They used radioactive sulfur (35S) to identify protein and radioactive phosphorus (32P) to identify the components of nucleic acid.
3. The tadpole-shaped bacteriophage attaches to the bacteria. Its genetic material enters the bacterial cell by dissolving the cell wall of bacteria. The bacterial cell treats the viral genetic material as if it was it’s own and subsequently manufactures more virus particles.
4. Infection: They grew some viruses on a medium that contained radioactive phosphorus and others on a medium that contained radioactive sulfur.
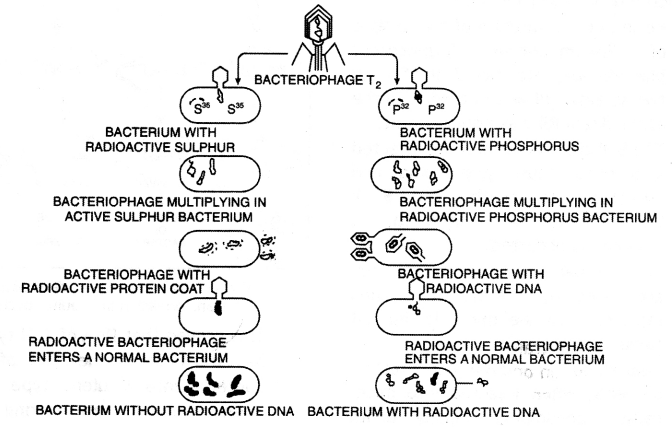
The Hershey—Chase Experiment
5. Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive protein because DNA contains phosphorus but protein does not. Similarly, viruses grown on radioactive sulfur contained radioactive protein but not radioactive DNA because DNA does not contain sulfur.
6. Radioactive phages were allowed to attach to E. co/i bacteria. Then as the infection proceeded, the viral coats were removed from the bacteria by agitating them in a blender. The virus particles were separated from the bacteria by spinning them in a centrifuge.
7. Bacteria that were infected with viruses that had radioactive DNA were radioactive, indicating that DNA was the material that passed from the virus to the bacteria.
8. Bacteria that were infected with viruses that had radioactive proteins were not radioactive.
9. This indicates that proteins did not enter the bacteria from the viruses.
10. DNA is, therefore, the genetic material that is passed from virus to bacteria.
Question 23.
List the characteristics of DNA molecules.
Answer:
Characteristics of DNA molecule:
- The two chains are spirally coiled about around a common axis to form a regular, right-handed double helix.
- The double helix has a major groove and minor groove alternately.
- The helix is 20Å wide; it’s one complete turn is 34Å long, and has 10 base pairs, and the successive base pairs are 3.4Å apart.
- The two chains are complementary to each other with respect to base sequence.
- The two strands are hydrogen-bonded: A on one chain is joined to T on the other chain by 2 hydrogen
bonds; C on one chain is linked to G on the other chain by 3 hydrogen bonds. - The two strands run in an antiparallel direction.
- The amount of A + G = the amount of T + C; the amount of A = the amount of T: the amount of G = the amount of C. Sugar and phosphate groups occur in equal proportion.
- The DNA molecule is remarkably stable due to hydrogen bonding and hydrophobic interactions.
- The DNA molecule undergoes denaturation and renaturation easily.
- The denatured DNA strands are hyperchromic.
- The DNA molecule can replicate and repair itself, and can also transcribe RNAs.
- The base sequence of one chain serves as the genetic code.
- The DNA can function in vitro.
- The amount of DNA per nucleus is constant in all the body cells of a given species.
Question 24.
How is a long DNA molecule adjusted in a nucleus? (CBSE Delhi 2008)
Or
(i) What is this diagram representing?
(ii) Name the parts A, B, and C.
(iii) In the eukaryotes, the DNA molecules are organized within the nucleus. How is the DNA molecule organized in a bacterial cell in the ab¬sence of a nucleus? (CBSE 2009)
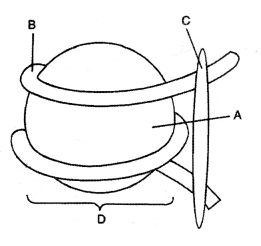
Answer:
The distance between two consecutive base pairs is 0.34 nm (0.34 × 10-9 m). If the length of DNA double helix in a typical mammalian cell is calculated (simply by multiplying the total number of bp with the distance between two consecutive bp, that is, 6.6 × 109 bp × 0.34 × 1(T9 m/bp), it comes out to be approximately 2.2 meters. A length that is far greater than a dimension of a typical nucleus (approx 10-6 m).
In eukaryotes, this organization is much more complex. There is a set of positively charged, basic proteins called ‘histones’. A protein acquires charge depending upon the abundance of amino acid residues with charged side chains. Histones are rich in amino acid residues lysines and arginines. Both the amino acid residues carry positive charges in their side chains. Histone organized to form a unit of eight histone molecules called ‘histone octamer’. The negatively charged DNA is wrapped around positively charged histone-octamer to form a structure called ‘Nucleosome’.
A typical nucleosome contains 200 bp of DNA helix. Nucleosomes constitute the repeating unit of a structure in the nucleus called ‘Chromatin’, thread-like stained (colored) bodies seen in the nucleus. The nucleosomes in chromatin are seen as a “beads-on-string” structure when viewed under an electron microscope (EM).
Or
(i) Nucleosome
(ii)
(iii) Organization of DNA molecule in bacteria. In prokaryotes like bacteria which lacks a nucleus, DNA is not scattered. The negatively charged DNA binds with positively charged proteins in a region called a nucleoid.
Question 25.
Describe briefly the mechanism of DNA replication. (CBSE (Delhi) 2019)
Or
Draw a labeled diagram of the replication fork. (CBSE 2011)
Or
Draw a neat labeled sketch of replicating fork of DNA.
Or
(a) Why does DNA replication occur within a replication fork and not in its entire length simultaneously?
(b) “DNA replication is continuous and discontinues on the two strands within the replication fork.” Give reasons. (CBSE (Outside Delhi) 2019)
Answer:
Mechanism of DNA replication. Watson and Crick, while explaining their model of DNA structure, suggested the semi-conservative mechanism of its replication. The quality and quantity of DNA in parent and daughter cells must be the same.
Replication is one of the most important properties of DNA and forms the very basis of life:
1. Replication takes place during the S-phase of the interphase between two mitotic cycles.
2. Replication is a semi-conservative process in which each of the two double helices formed from the parent double strand has one old and one new strand. Repair replication is non-conservative.
3. DNA replication requires a DNA template, a primer, deoxyribonucleoside triphosphates (dATP, dGTP, dTTP, dCTP), Mg++, DNA unwinding protein, superhelix relaxing protein, a modified RNA polymerase to synthesize the RNA primer, a joining polynucleotide ligase, enzyme.
4. Watson and Crick suggested that the two strands of DNA molecules uncoil and separate, and each strand serves as a template for the synthesis of a new strand alongside it.
5. The sequence of bases which should be present in the new strands can be easily predicted because these would be complementary to the bases present in the old strands. A will pair with T, T with A, C with G, and G with C.
6. Thus, two daughter molecules are formed from the parent molecule and these are identical to the parent molecule.
7. Each daughter DNA molecule consists of one old (parent) strand and one new strand.
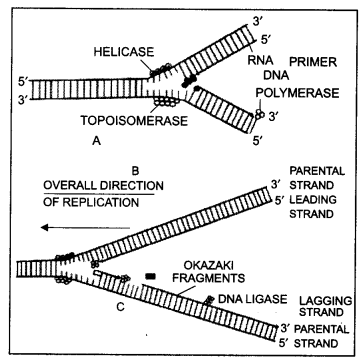 Continuous and discontinuous synthesis of DNA.
Continuous and discontinuous synthesis of DNA.
8. Since only one parent strand is conserved in each daughter molecule, this mode of replication is said to be semiconservative.
9. In one strand replication takes place discontinuously and short pieces called Okazaki fragments are synthesized. One strand may synthesize a continuous strand and the other Okazaki fragments. Both new strands are synthesized in the 5’ → 3’ direction. Thus one strand is synthesized forward and the other backward.
10. The Okazaki pieces are joined by polynucleotide ligase, a joining enzyme, to form continuous strands.
Question 26.
Provide experimental evidence for the semi-conservative mode of replication of DNA. (CBSE 2008 (Outside Delhi) 2012, 2016, 2019 C)
Answer:
Meselson and Stahl (1958) experimentally proved that DNA replication is semi-conservative.
- E. coli bacterium was grown for many generations in a culture medium in which the nitrogen source contained heavy isotope N15 thus the labeling of bacterial DNA was done.
- Later on, these bacteria were cultured in N14 non-radioactive isotope.
- DNA was analyzed to determine the distribution of radioactivity.
- The experiment showed that one strand of each daughter DNA molecule was radioactive whereas the other was non-radioactive.
- During the second replication in the N14 medium, the radioactive and non-radioactive strands separated and served as the template for the synthesis of non-radioactive strands.
- Out of four DNA molecules, two are completely non-radioactive and the other two have half of the molecule as non-radioactive.
- This evidence shows that DNA replication is semi-conservative.
- This biochemical evidence was supported by the direct cytological observation of duplicating DNA of E. coli.
Question 27.
(i) State the ‘Central dogma’ as proposed by Francis Crick. Are there any exceptions to it? Support your answer with a reason and an example.
Answer:
Francis Crick proposed the Central dogma in molecular biology, which states that the genetic information
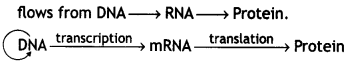
Central dogma
In some viruses, central dogma is seen in the reverse direction, that is from RNA to DNA as RNA is the main genetic material. Example: Retrovirus (HIV) and the process is reverse transcription.
(ii) Explain how the biochemical characterization (nature) of the ‘Transforming Principle’ was determined, which was not defined from Griffith’s experiments. (CBSE Delhi 2018)
Answer:
Transforming principle: In 1928, Frederick Griffith, in a series of experiments with Streptococcus pneumonia which is a bacterium responsible for pneumonia, witnessed a miraculous transformation in the bacteria. During the experiment, a living organism (bacteria) changed its physical form. He concluded that the R strain bacteria had somehow been transformed by the heat-killed S strain bacteria.
Some ‘transforming principle’, transferred from the heat-killed S strain, had enabled the R strain to synthesize a smooth polysaccharide coat and become virulent. This must be due to the transfer of the genetic material. However, the biochemical nature of genetic material was not defined from his experiments.
Oswald Avery, Colin MacLeod, and Maclyn McCarty worked to determine the biochemical nature of the ‘transforming principle’ in Griffith’s experiment. They purified biochemicals (proteins, DNA, RNA, etc.) from the heat-killed S cells to see which ones could transform live R cells into S cells. They discovered that DNA alone from S bacteria caused R bacteria to become transformed.
They also discovered that protein-digesting enzymes (proteases) and RNA-digesting enzymes (RNases) did not affect transformation, so the transforming substance was not a protein or RNA. Digestion with DNase did inhibit transformation, suggesting that the DNA caused the transformation. They concluded that DNA is the hereditary material, but not all biologists were convinced.
Question 28.
(a) Write the contributions of the following scientists in deciphering the genetic code: George Gamow; Hargobind Khorana; Marshall Nirenberg; Severo Ochoa.
Answer:
Contributions of scientists:
- George Gamow: He suggested that in order to code for all the 20 Amino acids, the code should be made up of three nucleotides. He proved that the codon is a triplet.
- Hargobind Khorana: He provided experimental proof that genetic codon is always triplet. He was able to synthesize RNA molecules with a defined combination of bases (homopolymers and copolymers)
- Marshall Nirenberg: He prepared a cell-free system for protein synthesis and finally deciphered the genetic code.
- Severe Ochoa: He established that enzyme polynucleotide phosphorylase was helpful in RNA synthesis with a defined sequence in a template-independent manner.
(b) State the importance of a Genetic code in protein biosynthesis. (CBSE Delhi 2019)
Answer:
Importance of Genetic code: Genetic code codes for a specific amino acid that is required for protein synthesis. (CBSE Outside Delhi 2018)
Question 29.
Explain translation in detail.
Or
Explain the process of charging tRNA.
Answer:
Translation: During the translation process, proteins are made by the ribosomes on the mRNA strand:
The main steps are:
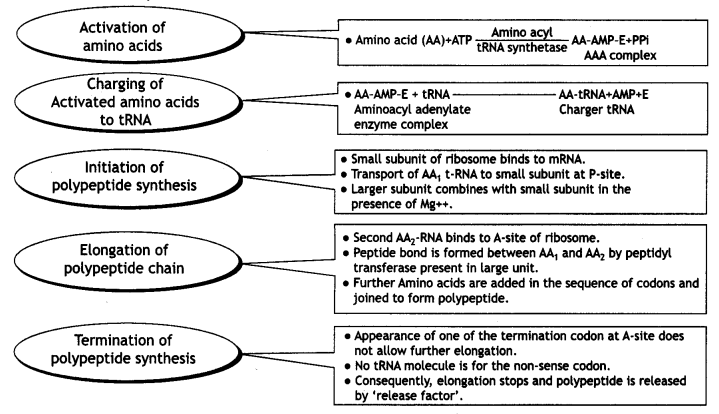
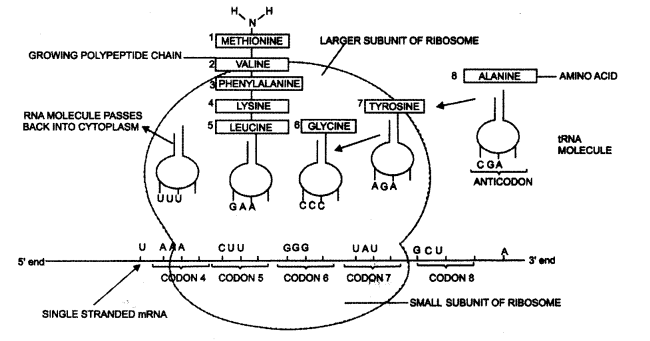
Continuous and discontinuous synthesis of DNA:
- Activation of amino acid.
- Transfer of activated amino acid to tRNA.
- Initiation of synthesis.
- Elongation of a polypeptide chain.
- Termination of a chain.
Question 30.
Explain the steps involved in DNA fingerprinting. (CBSE 2009)
Answer:
Steps of DNA fingerprinting:
The steps/procedure in DNA fingerprinting include the following:
- Extraction: DNA is extracted from the cells in a high-speed, refrigerated centrifuge.
- Amplification: Many copies of the extracted DNA are made by a polymerase chain reaction.
- Restriction Digestion: DNA is cut into fragments with restriction enzymes into precise reproducible sequences.
- Separation of DNA sequences/ restriction fragments: The cut DNA fragments are introduced and passed through an electrophoresis setup containing agarose polymer gel; the separated fragments can be visualized by staining them with a dye that shows fluorescence under ultraviolet radiation.
Lac operon - Southern Blotting: The separated DNA sequences are transferred onto a nitrocellulose or nylon membrane.
- Hybridization: The nylon membrane is immersed in a bath and radioactive probes (DNA segments of known sequence) are added: these probes target a specific nucleotide sequence that is complementary to them.
- Autoradiography: The nylon membrane is pressed on an X-ray film and dark bands develop at the probe sites.
Question 31.
What is the inducer in the lac operon? How does it ensure the “switching on” of genes?
(i) Draw a schematic representation of the Lac operon.
Answer:
Inducer. It is a chemical that may be a substrate, hormone, or some other metabolite which after coming in contact with the repressor, changes the latter into the non-DNA binding state so as to free the operator gene. Thus the “switch on” occurs.
(ii) Explain how does this operon gets switched ‘on’ or ‘off’.
Answer:
The expression of the genes is usually controlled to achieve maximum cellular economy. This means that the gene will be turned on or off as per requirement. A set of genes will be switched on when there is the necessity to handle and metabolize a new substrate. When these genes are turned on enzymes are produced, which metabolize the new substrate. The phenomenon is known as induction.
(iii) What is the role of a regulatory gene? (CBSE Delhi 2012, 2019 C)
Answer:
Role of the regulatory gene in a lac operon: It synthesizes a biochemical or regulator protein that can act positively as an activator and negatively as a repressor. It controls the activity of the operator gene.
Question 32.
(i) Describe the structure and function of a tRNA molecule. Why is it referred to as an adapter molecule?
Answer:
t-RNA (transfer RNA) reads the genetic code on one hand and transfers amino acids on the other hand. So it was called adapter molecule by Francis Crick. It is also called soluble RNA (SRNA).
Note: For the figure, please see the answer of Q. no. 11 short answer type question. The secondary structure of tRNA is clover leaf-like but the 3-D structure is inverted L-shaped. t-RNA has five arms or loops:
- Anticodon loop: It has bases complementary to the code.
- Amino acid acceptor end: Amino acid binds to it.
- T-loop: It helps in binding to ribosomes.
- D-loop: It helps in binding aminoacyl synthetase.
- Variable loop: Its function is not known.
(ii) Explain the process of splicing of hn-RNA in a eukaryotic cell. (CBSE Delhi 2017)
Answer:
The primary transcript formed in eukaryotes is non-functional, containing both the coding region exon and non-coding region intron in RNA and are called heterogeneous RNA or hn-RNA. hn-RNA undergoes a process where the introns are removed and exons are joined to form mRNA by the process called splicing.
Question 33.
(i) Why does replication occur in small replication forks and not in entire lengths?
Answer:
Replication occurs in a small opening of a DNA helix called replication forks for very long DNA molecules. The reason is in the case of long DNA molecules, the two strands of DNA cannot be separated in their entire length due to very high energy demand.
(ii) Why is DNA replication continuous and discontinuous in a replication fork?
Answer:
DNA-dependent DNA polymerase catalyzes DNA polymerization only in one direction, that is, 5′ → 3′. DNA strands are anti-parallel and have opposite polarity. So on template strand with polarity 3′ → 5′ DNA replication is continuous white on the template strand with polarity 5′ → 3′ replication is discontinuous.
(iii) State the importance of the origin of replication in a replication fork. (CBSE Delhi 2018C)
Answer:
Replication cannot start randomly at any point in DNA. It requires a definite region of sequences where the replication originates. This site is called the origin of replication.
Question 34.
Compare the processes of DNA replication and transcription in prokaryotes. (CBSE Delhi 2019C)
Answer:
| Replication | Transcription |
| (i) It is a synthesis of DNA from DNA. | (i) It is a synthesis of RNA from DNA. |
| (ii) Both strands take part in replication. | (ii) Only one strand functions as a template. |
| (iii) It forms double-stranded DNA. | (iii) It forms single-stranded RNA. |
| (iv) RNA primer is essential for initiation. | (iv) No primer Is required. |
| (v) Occurs in the S phase of the cell cycle. | (v) Occurs in the G1 and G2 phases of the cell cycle. |
| (vi) Catalysed by DNA polymerase enzymes. | (vi) Catalysed by RNA polymerase enzymes. |
| (vii) Deoxyribonucteoside triphosphates (dATP, dGTP, dCTP, dTTP) serve as raw materials. | (vii) Ribonucleoside triphosphates (ATP, GTP, CTP, UTP) serve as raw materials. |
| (viii)Involves unwinding and splitting of the entire DNA molecule (chromosome). | (viii) Involves unwinding and splitting of only those genes which are to be transcribed. |
| (ix) Two double-stranded DNA molecules are formed from one DNA molecule. | (ix) A single one-strand RNA molecule is formed from a segment of one DNA strand. |
Question 35.
Write the different components of a lac operon in E. coil. Explain its expression while in an ‘open’ state. (CBSE Delhi 2017, 2019 C)
Answer:
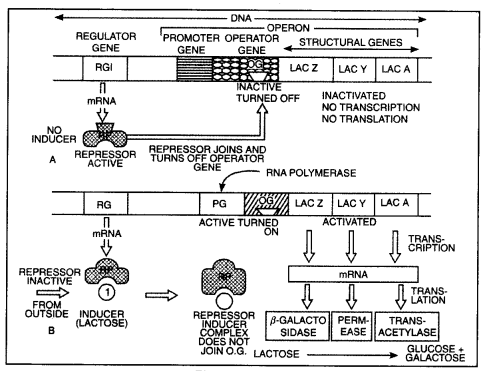
The Lac operon (Inducible operon): The concept of the operon was first proposed in 1961 by Jacob and Monod.
Components of an operon:
- Structural genes: It Is the fragment of DNA that transcribes mRNA for polypeptide synthesis.
- Promoter: It Is the sequence of DNA where RNA poLymerase binds and initiates transcription.
- Operator: It is the sequence of DNA adjacent to the promoter.
- Regulator gene: It is the gene that codes for repressor protein which binds to the operator due to which operon Is switched “off”.
- Inducer: Lactose Is an Inducer that helps in switching “on” of an operon. Lac operon consists of three structural genes (z, y, a), operator (o), promoter (p), regulatory gene (r).
(a)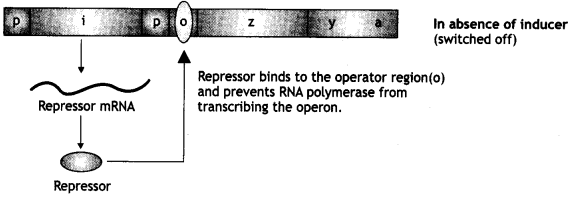
(b) 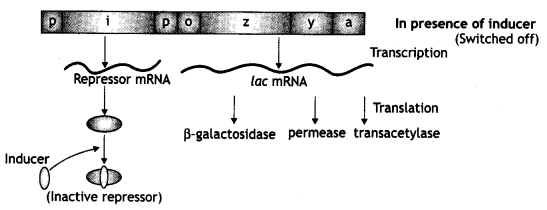
- Gene z codes for p-galactosidase
- Gene y codes for permease.
- Gene codes for enzymes transacetylase. When lactose is absent:
When lactose is absent, i.e. gene produces repressor protein.
This repressor protein binds to the operator and as a result, prevents RNA polymerase to bind to the operon, and the operon is switched off.
When lactose is present:
- Lactose acts as an inducer that binds to the repressor and forms an inactive repressor.
- The repressor cannot bind to the operator.
- Now the RNA polymerase binds to the operator and transcribes lac mRNA.
- Lac mRNA is polycistronic, i.e. produces all three enzymes p-galactosidase, permease, and transacetylase.
- The lac operon is switched on.
Question 36.
What is the human genome project (HGP)? Write salient features of the human genome project.
Answer:
Human Genome Project. It is a mega project involving a lot of money, the most advanced techniques, numerous computers, and scientists at work. The magnitude of the project can be imagined that if the cost of sequencing a bp is 3 dollars, sequencing of 3 × 109 bp would be a billion dollars. If the data is to be stored in books, with each book having 1000 pages and each page with 1000 letters, some 3300 books will be required. Here bioinformatics databasing and other high-speed computational devices have helped in the analysis, storage, and retrieval of information.
Salient features of the human genome:
- The human genome contains 3164.7 million nucleotides (base pairs).
- The size of the genes varies; an average gene consists of 3000 bases, while the largest gene, dystrophin, consists of 2.4 million bases.
- The total number of genes is estimated to be 30000 and 99.9% of the nucleotides are the same in humans.
- The functions of over 50% of the discovered genes are not known.
- Only less than 2% of the genome codes for proteins.
- Repetitive segments made up a large portion of the human genome.
- Repetitive sequences throw light on chromosome structure and dynamics and evolution, though they are thought to have no direct coding functions.
- (Chromosome 1 has 2968 genes and Y-chromosome has the least number (231 genes).
- Scientists have identified about 1.4 million locations, where DNA differs in a single base in human beings. These are called single nucleotide polymorphisms (SNPs).
- Repeated sequences make up a large portion of the human genome.
Very Important Figures:
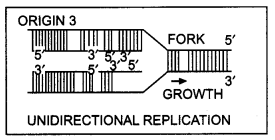 Unidirectional repLication
Unidirectional repLication
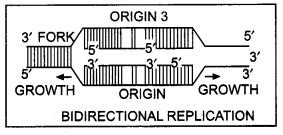 BidirectionaL replication
BidirectionaL replication
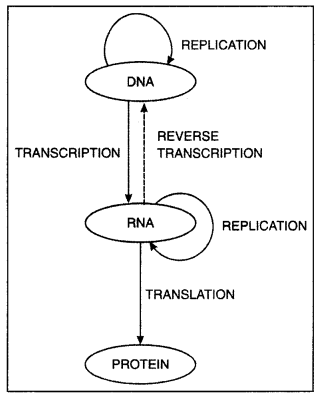 The flow of genetic information.
The flow of genetic information.
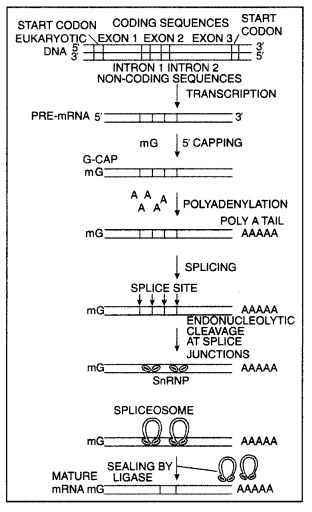 Transcription in eukaryotes
Transcription in eukaryotes