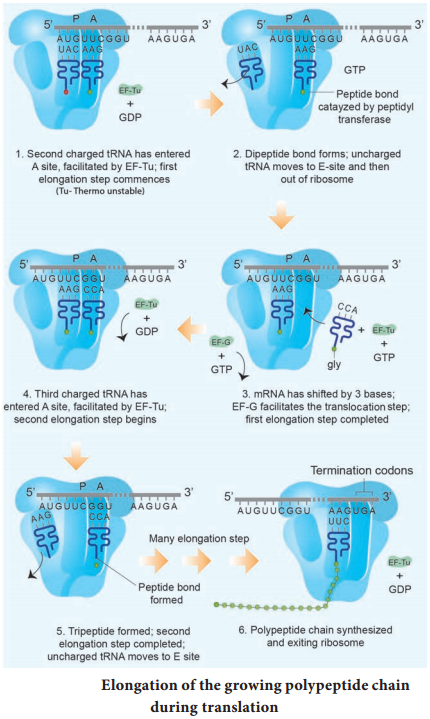
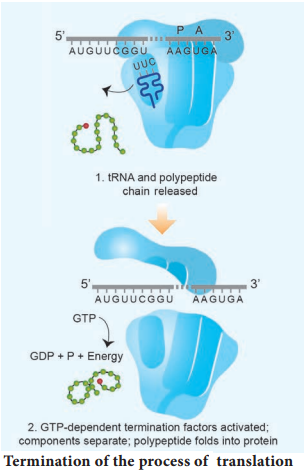
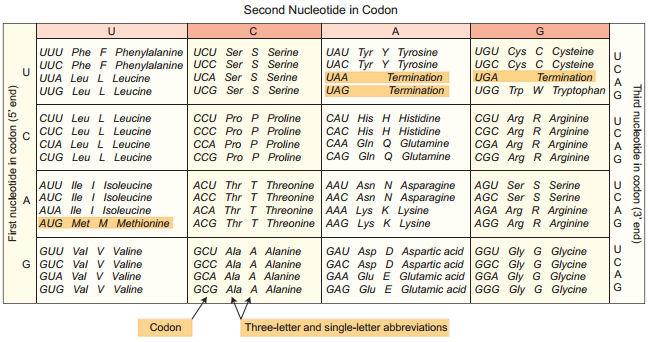
Learninsta presents the core concepts of Biology with high-quality research papers and topical review articles.
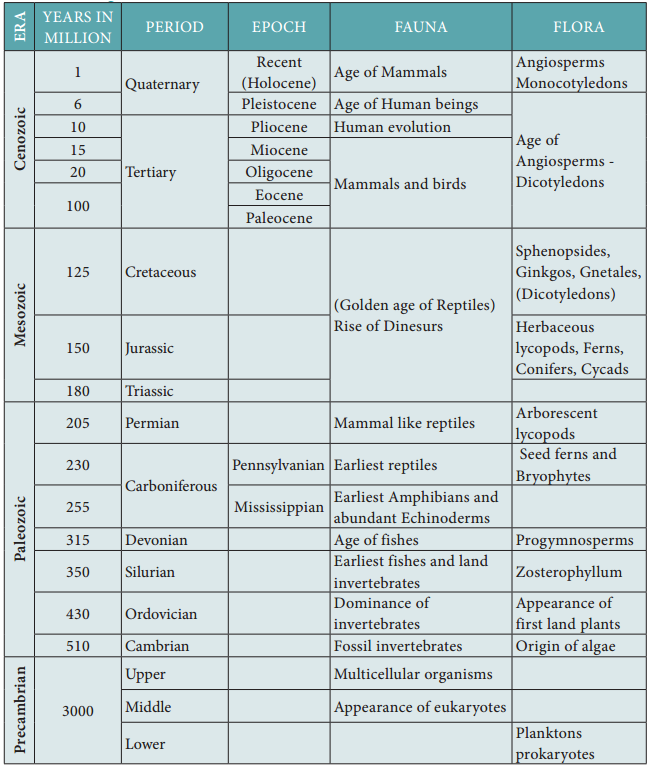
Theories Of Biological Evolution
Lamarck’s theory
Jean Baptiste de Lamarck, was the first to postulate the theory of evolution in his famous book ‘Philosophie Zoologique’ in the year 1809. The two principles of Lamarckian theory are:
(i) The theory of use and disuse – Organs that are used often will increase in size and those that are not used will degenerate. Neck in giraffe is an example of use and absence of limbs in snakes is an example for disuse theory.
(ii) The theory of inheritance of acquired characters – Characters that are developed during the life time of an organism are called acquired characters and these are then inherited.
The main objection to Lamarckism
Lamarck’s “Thory of Acquired characters” was disproved by August Weismann who conducted experiments on mice for twenty generations by cutting their tails and breeding them. All mice born were with tail. Weismann proved his germplasm theory that change in the somatoplasm will not be transferred to the next generation but changes in the germplasm will be inherited.
Neo-Lamarckism
The followers of Lamarck (Neo-Lamarckists) like Cope, Osborn, Packard and Spencer tried to explain Lamarck’s theory on a more scientific basis. They considered that adaptations are universal. Organisms acquire new structures due to their adaptations to the changed environmental conditions. They argued that external conditions stimulate the somatic cells to produce certain ‘secretions’ which reach the sex cells through the blood and bring about variations in the offspring.
Darwin’s theory of Natural Selection
Charles Darwin explained the theory of evolution in his book ‘The Origin of Species by Natural Selection’. During his journey around the Earth, he made extensive observations of plants and animals. He noted a huge variety and remarkable similarities among organisms and their adaptive features to cope up to their environment. He proved that fitest organisms can survive and leave more progenies than the unfit ones through natural selection.
Darwin’s theory was based on several facts, observations and influences. They are:
1. Over production (or) prodigality of production:
All living organisms increase their population in larger number. For example, Salmon fish produces about 28 million eggs during breeding season and if all of them hatch, the seas would be filed with salmon in few generations. Elephant, the slowest breeder that can produce six young ones in its life time can produce 6 million descendants at the end of 750 years in the absence of any check.
2. Struggle for existence:
Organisms struggle for food, space and mate. As these become a limiting factor, competition exists among the members of the population. Darwin denoted struggle for existence in three ways – Intra specific struggle between the same species for food, space and mate. Inter specific struggle with different species for food and space. Struggle with the environment to cope with the climatic variations, flood, earthquakes, drought, etc.,
3. Universal occurrence of variations
No two individuals are alike. There are variations even in identical twins. Even the children born of the same parents differ in colour, height, behavior, etc., The useful variations found in an organism help them to overcome struggle and such variations are passed on to the next generation.
4. Origin of species by Natural Selection
According to Darwin, nature is the most powerful selective force. He compared origin of species by natural selection to a small isolated group. Darwin believed that the struggle for existence resulted in the survival of the fittest. Such organisms become better adapted to the changed environment.
Objections to Darwinism
Some objections raised against Darwinism were –
- Darwin failed to explain the mechanism of variation.
- Darwinism explains the survival of the fittest but not the arrival of the fittest.
- He focused on small fluctuating variations that are mostly non-heritable.
- He did not distinguish between somatic and germinal variations.
- He could not explain the occurrence of vestigial organs, over specialization of some organs like large tusks in extinct mammoths, oversized antlers in the extinct Irish deer, etc.,
Neo Darwinism
Neo Darwinism is the interpretation of Darwinian evolution through Natural Selection as it has been modified since it was proposed. New facts and discoveries about evolution have led to modifications of Darwinism and is supported by Wallace, Heinrich, Haeckel, Weismann and Mendel. This theory emphasizes the change in the frequency of genes in population arises due to mutation, variation, isolation and Natural
selection.
Mutation theory
Hugo de Vries put forth the Mutation theory. Mutations are sudden random changes that occur in an organism that is not heritable. De Vries carried out his experiments in the Evening Primrose plant (Oenothera lamarckiana) and observed variations in them due to mutation.
According to de Vries, sudden and large variations were responsible for the origin of new species whereas Lamarck and Darwin believed in gradual accumulation of all variations as the causative factors in the origin of new species. Hugo de Vries believed that Mutations are random and directionless, but Darwinian variations are small and directional.
Salient features of Mutation Theory
- Mutations or discontinuous variation are transmitted to other generations.
- In naturally breeding populations, mutations occur from time to time.
- There are no intermediate forms, as they are fully fledged.
- They are strictly subjected to natural selection.
Modern synthetic theory
Sewell Wright, Fisher, Mayer, Huxley, Dobzhansky, Simpson and Haeckel explained Natural Selection in the light of Post-Darwinian discoveries. According to this theory gene mutations, chromosomal mutations, genetic recombinations, natural selection and reproductive isolation are the fie basic factors involved in the process of organic evolution.
(i) Gene mutation
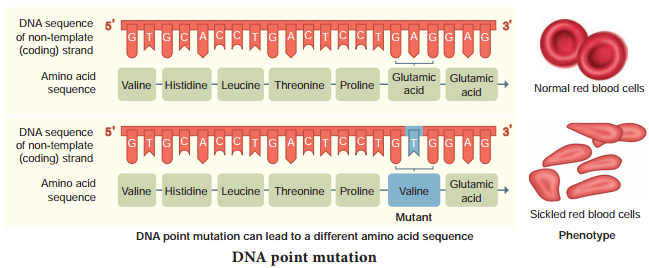
Refers to the changes in the structure of the gene. It is also called gene/point mutation. It alters the phenotype of an organism and produces variations in their offspring.
(ii) Chromosomal mutation
Refers to the changes in the structure of chromosomes due to deletion, addition, duplication, inversion or translocation. This too alters the phenotype of an organism and produces variations in their offspring.
(iii) Genetic recombination
Is due to crossing over of genes during meiosis. This brings about genetic variations in the individuals of the same species and leads to heritable variations.
(iv) Natural selection
Does not produce any genetic variations but once such variations occur it favours some genetic changes while rejecting others (driving force of evolution).
(v) Reproductive Isolation
Helps in preventing interbreeding between related organisms.
Evolution by anthropogenic sources
Natural Selection (Industrial melanism)
Natural selection can be explained clearly through industrial melanism. Industrial melanism is a classical case of Natural selection exhibited by the peppered month, Biston betularia. These were available in two colours, white and black. Before industrialization peppered moth both white and black coloured were common in England. Pre-industrialization witnessed white coloured background of the wall of the buildings hence the white coloured months escaped from their predators.
Post industrialization, the tree trunks became dark due to smoke and soot let out from the industries. The black moths camouflaged on the dark bark of the trees and the white moths were easily identified by their predators. Hence the dark coloured month population was selected and their number increased when compared to the white months. Nature offered positive selection pressure to the black coloured months. The above proof shows that in a population, organisms that can adapt will survive and produce more progenies
resulting in increase in population through natural selection.
Artificial selection is a byproduct of human exploitation of forests, oceans and fisheries or the use of pesticides, herbicides or drugs. For hundreds of years humans have selected various types of dogs, all of which are variants of the single species of dog. If human beings can produce new varieties in short period, then “nature” with its vast resources and long duration can easily produce new species by selection.
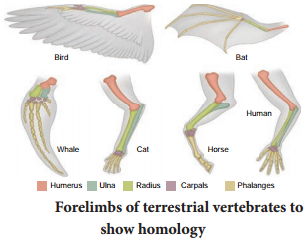
Adaptive Radiation
The evolutionary process which produces new species diverged from a single ancestral form becomes adapted to newly invaded habitats is called adaptive radiation. Adaptive radiations are best exemplified in closely related groups that have evolved in relatively short time.
Darwin’s finches and Australian marsupials are best examples for adaptive radiation. When more than one adaptive radiation occurs in an isolated geographical area, having the same structural and functional similarity is referred to as convergent evolution.
Darwin’s fiches
Their common ancestor arrived on the Galapagos about 2 million years ago. During that time, Darwin’s finches have evolved into 14 recognized species differing in body size, beak shape and feeding behavior. Changes in the size and form of the beak have enabled different species to utilize different food resources such as insects, seeds, nectar from cactus flowers and blood from iguanas, all driven by Natural selection. Fig. 6.5 represents some of the finches observed by Darwin.
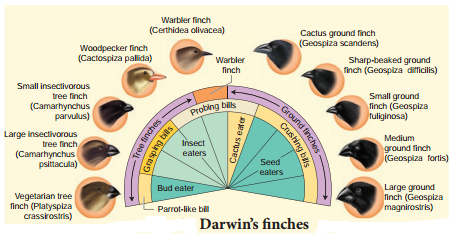
Genetic variation in the ALX1 gene in the DNA of Darwin finches is associated with variation in the beak shape. Mild mutation in the ALX1 gene leads to phenotypic change in the shape of the beak of the Darwin finches. Marsupials in Australia and placental mammals in North America are two subclasses of mammals they have adapted in similar way to a particular food resource, locomotory skill or climate.
They were separated from the common ancestor more than 100 million year ago and each lineage continued to evolve independently. Despite temporal and geographical separation, marsupials in Australia and placental mammals in North America have produced varieties of species living in similar habitats with similar ways of life. Their overall resemblance in shape, locomotory mode, feeding and foraging are superimposed upon different modes of reproduction. This feature reflects their distinctive evolutionary relationships.
Over 200 species of marsupials live in Australia along with many fewer species of placental mammals. The marsupials have undergone adaptive radiation to occupy the diverse habitats in Australia, just as the placental mammals have radiated across North America.