Here we are providing Class 12 Biology Important Extra Questions and Answers Chapter 11 Biotechnology: Principles and Processes. Important Questions for Class 12 Biology are the best resource for students which helps in Class 12 board exams.
Class 12 Biology Chapter 11 Important Extra Questions Biotechnology: Principles and Processes
Biotechnology: Principles and Processes Important Extra Questions Very Short Answer Type
Question 1.
What is genetic engineering?
Answer:
Genetic engineering. It is a technique for artificially and deliberately modifying DNA (genes) to suit human needs. It is also called recombinant DNA technology or DNA splicing. It is a kind of biotechnology.
Question 2.
Define recombinant DNA.
Answer:
Recombinant DNA. They are molecules of DNA that are formed through genetic recombination methods.
Question 3.
What is the role of restriction endonuclease?
Answer:
Restriction endonucleases are specific enzymes which can cleave double-stranded DNA at the specific site.
Question 4.
What are BACs and YACs? (CBSE 2016)
Answer:
BACs and YACs are artificial chromosomes from bacteria and yeast efficient for gene transfer. They are vectors.
Question 5.
Name the soil bacterium which contains the gene for production of endotoxins.
Answer:
Agrobacterium tumefaciens.
Question 6.
Name a technique by which DNA fragments can be separated. (CBSE Delhi 2008)
Answer:
Gel electrophoresis.
Question 7.
What is the principle of Gel electrophoresis?
Answer:
DNA fragments are negatively charged so they move to anode under electric field through the matrix (usually agarose). This matrix gel acts as sieve and DNA fragments resolve according to their size.
Question 8.
Name the compound used for staining DNA to be used in Recombinant Technology. What is the colour of such stained DNA?
Answer:
The compound used for staining DNA is ethidium bromide. Stained DNA becomes orange.
Question 9.
Name the technique for vector less direct gene transfer.
Answer:
Gene gun.
Question 10.
What is the role of ‘Ori’ in any plasmid?
Answer:
The plasmid is prokaryotic circular DNA which has a sequence of nucleotides from where the replication starts. This is called the origin of replication.
Role of Ori is to start replication. Also, the copy number of linked DNA is controlled by Ori.
Question 11.
Do normal £. coli cells have any gene resistance against antibiotics?
Answer:
No.
Question 12.
What is the function of TPA?
Answer:
TPA (Tissue plasminogen activator) dissolves blood clots after a heart attack and stroke.
Question 13.
Give an example in which recombinant DNA technology has provided a broad range of tool in the diagnosis of diseases.
Answer:
Construction of probes, which are short segments of single-stranded DNA attached to a radioactive or fluorescent marker.
Question 14.
Give the full form of PCR. Who developed it? (CBSE Delhi 2013)
Answer:
PCR is a polymerase chain reaction. It was developed by Kary Mullis in 1985.
Question 15.
What is the source of DNA polymerase, i.e. Taq polymerase? (CBSE Outside Delhi 2013)
Answer:
Taq polymerase is isolated from the bacterium Thermus Aquaticus.
Question 16.
Define “melting of target DNA”.
Answer:
The target DNA containing the sequence to be amplified is heat-denatured (around 94° C for 15 seconds) to separate its complementary strands. This process is called melting of target DNA.
Question 17.
Expand ELISA. Write one application. (CBSE Delhi 2013)
Answer:
ELISA-Enzyme Linked ImmunoSorbent Essay. Importance-lt is used for the diagnosis of AIDS.
Question 18.
What are transgenic animals? Give one example. (CBSE Outside Delhi 2016)
Answer:
Transgenic animals: The animals obtained by genetic engineering containing transgenes are known as transgenic animals.
Example. Transgenic cow ‘Rosie’.
Question 19.
How many PCR cycles are adequate for proper amplification of DNA segment?
Answer:
20-30 cycles.
Question 20.
Define gene therapy.
Answer:
Gene Therapy: It is the replacement of a faulty gene by normal healthy functional gene.
Question 21.
What is the importance of gene bank?
Answer:
It provides a stock from which genes can be obtained for improving the varieties or used in genetic engineering.
Question 22.
What can be the source of thermostable DNA?
Answer:
Thermostable DNA is obtained from a bacterium Thermus Aquaticus.
Question 23.
What are selectable markers?
Answer:
Genes which are able to select transformed cell from the non-recombinant cells are called selectable markers.
Question 24.
Why is enzyme cellulase used for isolating genetic material from plant cells and not from animal cells? (CBSE 2010)
Answer:
Cellulase is used for breaking the cell wall of plant cells whereas animal cells lack a cell wall. The cell wall is made of cellulose which can be broken down by cellulase.
Question 25.
Give one example each of transgenic plant and transgenic animal.
Answer:
- Transgenic tomato plant called Flavor Savr.
- A transgenic mouse called Supermouse.
Question 26.
What would be the molar concentration of human DNA in a human cell?
Answer:
Humans have 3 M of DNA per cell, i.e. the molar concentration is 3.
Question 27.
Do eukaryotic cells have restriction endonucleases?
Answer:
Yes, eukaryotic cells possess restriction endonucleases. They are involved in editing (Proofreading) and DNA repairs during DNA replication.
Question 28.
Name a technique by which DNA fragments can be separated. (CBSE Delhi 2008)
Answer:
Get electrophoresis.
Question 29.
Biotechnological techniques can help to diagnose the pathogen much before the symptoms of the disease appear in the patient. Suggest any two such techniques. (CBSE Outside Delhi 2019)
Answer:
PCR – Polymerase chain reaction
ELISA – Enzyme-linked immunosorbent assay
Question 30.
Why is it not possible for an alien DNA to become part of chromosome anywhere along its length and replicate? (CBSE 2014)
Answer:
For multiplication of any alien DNA, it needs to be a part of a chromosome which has a specific sequence known as the origin of replication.
Question 31.
Mention the type of host cells suitable for the gene guns to introduce an alien DNA. (CBSE Delhi 2014)
Answer:
Plant cells.
Question 32.
Name the enzymes that are used for isolation of DNA from bacterial and fungal cells for rDNA technology. (CBSE 2014)
Answer:
Lysozyme for bacterial cell and chitinase for the fungal cell.
Question 33.
What is EcoRI? How does EcoRI differ from an exonuclease? (CBSE Outside Delhi 2015)
Answer:
EcoRI is an endonuclease restriction enzyme which cut both the stands of palindromic DNA at a specific position of nitrogen base 5′ (GAATTC) 3′ while exonuclease removes nucleotides from terminals of DNA strands.
Biotechnology: Principles and Processes Important Extra Questions Short Answer Type
Question 1.
(i) While cloning vectors, which of the two will be preferred by biotechnologists, bacteriophages or plasmids. Justify with reason.
Answer:
Biotechnologists prefer bacteriophages for cloning over plasmids because they have very high copy numbers of their genome within the bacterial cells whereas some plasmids may have only one or two copies per cell and others may have 15-100 copies per cell. Phage vectors are more efficient than plasmids for cloning large DNA fragments.
(ii) Name the first transgenic cow developed and state the Improvement In the quality of the product produced by it. (CBSE Sample paper 2018-19)
Answer:
Transgenic cow Rosie produced human protein-enriched milk (2.4 grams per litre).
Question 2.
What are the two core techniques that enabled the birth of biotechnology?
Answer:
The two core techniques that enabled the birth of modern biotechnology are:
- Genetic engineering techniques to alter the chemistry of genetic material (DNA and RNA), to introduce these into host organisms and thus change the phenotype of the host organism.
- Maintenance of sterile (microbial contamination-free) ambience in chemical engineering processes to enable the growth of only the desired microbe/eukaryotic cell in large quantities for the manufacture of biotechnological products like antibiotics, vaccines, enzymes, etc.
Question 3.
Make a list of tools of recombinant DNA technology. (CBSE Delhi, 2011)
Answer:
Key tools of recombinant DNA technology:
- Restriction enzymes
- Polymerase enzymes
- Ligases
- Vectors
- Host organism.
Question 4.
What does EcoRI signify? How its name is derived?
Answer:
EcoRI signifies the name of restriction endonuclease:
- First capital letter of the name that comes from the genus Escherichia is ‘E’.
- Second two small letters come from the species Coli of prokaryotic cells from which they are isolated, i.e. ‘co’.
- Letter R is derived from the name of the strain, i.e. Escherichia coli Ry 13.
- The Roman number indicates the order in which enzymes were isolated from that strain of bacteria.
Question 5.
What are recognition sequences or recognition sites?
Answer:
The sites recognised by restriction endonucleases are called recognition sites. The recognition sequences are different and specific for the different restriction endonucleases. These sequences are palindromic in nature.
Question 6.
Define vector. Give the properties of a “Good Vector”.
Answer:
A vector is a DNA molecule that has the ability to replicate in an appropriate host cell, and into which the DNA fragment to be cloned is integrated for cloning.
A good vector must have the following properties:
- It should have an origin of replication so that it is able to replicate autonomously.
- It should be easy to isolate and purify.
- It should get easily introduced into the host cells.
Question 7.
What is the difference between cloning and expression vectors?
Answer:
All vectors that are used for propagation of DNA inserts in a suitable host are called cloning vectors. When a vector is designed for the expression of, i.e. production of the protein specified by, the DNA insert, it is termed as an expression vector.
Question 8.
What do you understand by the term selectable marker?
Answer:
Selectable marker:
- A marker is a gene which helps in selecting those host cells which contain the vector (transformant) and eliminating the non-transformants. It selectively permits the growth of transformants.
- Common selectable markers for E. coli include the genes encoding resistance to antibiotics such as ampicillin. chloramphenicol tetracycline and kanamycin or the gene for (i-galactosidase which can be identified by a colour reaction. Normal E.Coli do not carry resistance against any of these antibodies.
Question 9.
Explain the principle that helps in separation of DNA fragments in Gel electrophoresis. (CBSE Delhi 2009 C)
Answer:
Get electrophoresis is a technique of molecules such as DNA/RNA/protein on the basis of their size under the influence of the electric field so that they migrate in the direction of electrode bearing the opposite charge. Positively charged molecules move towards cathode (-ve electrode) and vice versa. These molecules move through a medium or matrix and can be separated on the basis of their size.
Question 10.
Give the applications of PCR technology. (CBSE, Delhi 2013)
Answer:
- Amplification of DNA and RNA.
- Determination of orientation and location of restriction fragments relative to one another.
- Detection of genetic diseases such as sickle cell anaemia, phenylketonuria and muscular dystrophy.
Question 11.
Why is “Agrobacterium-mediated genetic transformation” in plants described as natural genetic engineer of plants? (CBSE Delhi 2011)
Answer:
Agrobacterium tumefaciens is a plant pathogenic bacterium which can transfer part of its plasmid DNA because it infects host plants. Agrobacterium produces crown gall in most of the dicotyledonous plants. These bacteria contain large tumours inducing plasmid (Ti-plasmids) which pass on their tumour causing gene into the genome of the host plant. Thus gene transfer is happening in nature without human involvement hence Agrobacterium-mediated genetic transformation is described as natural genetic engineering in plants.
Question 12.
Differentiate gene therapy and gene cloning.
Answer:
Differences between gene therapy and gene cloning:
| Gene therapy | Gene cloning |
| It is the replacement and/ or alteration of defective genes responsible for hereditary diseases by normal genes. | It is the technique of obtaining identical copies of a particular segment of DNA or a gene. |
Question 13.
From what you have learnt, can you tell whether enzymes are bigger or DNA is bigger in molecular size? How did you know?
Answer:
DNA molecules are bigger in size as compared to the molecular size of enzymes. Enzymes are proteins. Protein synthesis occurs from a small portion of DNA called genes.
Question 14.
How does restriction endonuclease work? (CBSE Delhi 2013, 2014, Outside Delhi 2019)
Or
What are molecular scissors? Explain their role. (CBSE 2009)
Answer:
Restriction endonuclease enzymes are called molecular scissors which can cut double-stranded DNA at specific sites.
Role of restriction endonuclease:
- Restriction endonuclease inspects the length of DNA sequence.
- It finds specific recognition sequence, i.e. palindromic nucleotide sequence in DNA.
- These enzymes cut the strand of DNA a little away from the centre of palindromic sites.
- Thus restriction endonucleases leave overhanging stretches called sticky ends on each strand.
Question 15.
How and why is the bacterium Thermus Aquaticus employed in recombinant DNA technology? Explain. (CBSE Delhi 2009)
Answer:
Thermus Aquaticus bacterium is employed in recombinant DNA technology because it has thermostable DNA polymerase (Taq. Polymerase) that remains active during high temperature-induced denaturation of a step of PCR.
This enzyme is employed during amplification of gene using PCR (Polymerase Chain Reaction). The amplified fragment can be used to ligate with a vector for further cloning.
Question 16.
Name the source of Taq polymerase. Explain its advantages. (CBSE Outside Delhi 2009)
Answer:
Taq Polymerase is extracted from Thermostable bacteria, namely Thermus Aquaticus. It remains active at a higher temperature and is used for denaturation of DNA during PCR.
Question 17.
What are recombinant proteins? How do bioreactors help in their production? (CBSE Outside Delhi 2009, 2015)
Answer:
Recombinant proteins. When any protein-encoding gene is expressed in a heterologous host, it is called recombinant protein. Bioreactors help in the production of recombinant proteins on large scale. A bioreactor provides optimal conditions for achieving the desired recombinant protein by biological methods.
Question 18.
What is meant by gene cloning?
Answer:
Formation of multiple copies of a particular gene is called gene cloning. A gene is separated and ligated to a vector-like plasmid. The recombinant plasmid is introduced into a plasmid-free bacterium through transformation. The transformed bacterium is made to multiply and form a colony. Each and every bacterium of the colony has a copy of the gene.
Question 19.
Both the wine marker and a molecular biologist who has developed a recombinant vaccine claim to be biotechnologist. Who in your opinion is correct?
Answer:
Both are considered biotechnologists. Wine marker utilises a strain of yeast which produces wine by fermentation. The molecular biologist uses a cloned gene for the antigen. The antigen is used as a vaccine. This permits the formation of antigen in huge quantity. Both generate products and services using living organisms useful to mankind.
Question 20.
You have created a recombinant DNA molecule by ligating a gene to a plasmid vector. By mistake, your friend adds an exonuclease enzyme to the tube containing the recombinant DNA. How will your experiment get affected as you plan to go to the transformation now?
Answer:
The experiment is not likely to be affected as the recombinant DNA molecule is circular and closed, with no free ends. Hence, it will not be a substrate for exonuclease enzyme which removes nucleotides from the free ends of DNA.
Question 21.
Explain the work carried out by Cohen and Boyer that contributed immensely to biotechnology. (CBSE2012)
Answer:
Work of Cohen and Boyer:
- Discovery of restriction endonuclease, an enzyme of E.coli which cut DNA at palindromic sequence.
- Preparation of recombinant DNA (Plasmid and DNA of interest.)
- Their work established recombinant DNA (rDNA) technology also called genetic engineering.
Question 22.
How are ‘Sticky ends’ formed on a DNA strand? Why are these so-called? (CBSE Delhi 2014)
Answer:
1. Restriction enzymes cut the strand of DNA a little away from the centre of palindrome site, but between the same two bases on opposite strands. As a result, single-stranded portions are left at each end. These overhanging stretches of DNA are called ‘Sticky ends’.
2. The Sticky ends are named so because they form hydrogen bonds with their complementary cut counterparts. The stickiness helps in the action of DNA ligase.
Question 23.
How is a continuous culture system maintained in bioreactors and why? (CBSE Delhi 2019)
Answer:
In order to maintain a continuous culture system, the used medium is drained out from one side of the bioreactor and the fresh medium is added from one side. This type of culturing method produces larger biomass leading to higher yields of the desired product.
Question 24.
Galactosidase enzyme is considered a better selectable marker. Justify the statement. (CBSE Delhi 2019)
Answer:
Recombinant strains can be differentiated from the non-recombinants ones easily by using this selectable marker. The selection is done on the basis of the colour change. All are grown on a chromogenic substance. Non-recombinants will change from colourless to blue while in recombinants insertional inactivation of galactosidase gene occurs.
Hence, recombinants showed no colour change. This is a single step, an easy method for selection.
Biotechnology: Principles and Processes Important Extra Questions Long Answer Type
Question 1.
What is genetic engineering? Explain briefly the distinct steps common to all genetic engineering technology.
Or
With the help of diagrams show the different steps in the formation of recombinant DNA by the action of restriction endonuclease. (CBSE 2011)
Answer:
Genetic engineering: It is a technique for artificially and deliberately modifying DNA (genes) to suit human needs. It is also called recombinant DNA technology or DNA splicing.
It is a kind of biotechnology:
- Isolation of genetic material which has the gene of interest.
- Cutting of gene of interest from genome and vector with the same restriction endonuclease enzyme. Amplifying gene of interest (PCR).
- Ligating gene of interest and vector using DNA ligase forming rDNA.
- Transformation of rDNA into the host cell.
- Multiplying host cell to create clones.
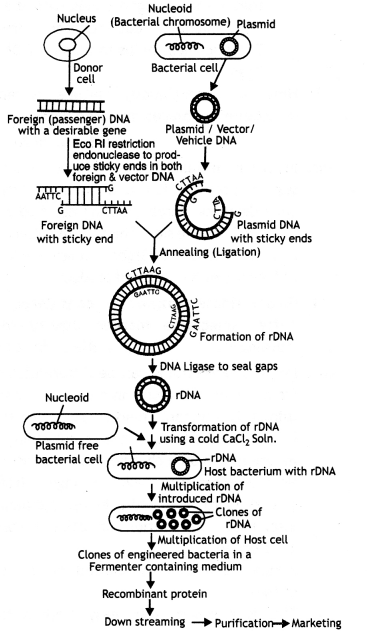
Diagram showing various steps Involved in DNA recombinant technology for the production of a recombinant protein.
Question 2.
List three important features necessary for preparing a genetically modifying organism.
Answer:
Conditions necessary for preparing:
- Identification of DNA with desirable genes.
- Introduction of the identified DNA into the host.
- Maintenance of introduced DNA in the host and transfer of the DNA to its progeny.
Question 3.
How are restriction endonuclease enzymes named? Write examples. (CBSE 2014
Answer:
The naming of restriction enzymes is as follows:
- The first letter of the name comes from the genus and the next two letters from the name of the species of the prokaryotic cell from which they are isolated.
- The next letter comes from the strain of the prokaryote.
- The roman numbers following these four letters indicate the order in which the enzymes were isolated from that strain of the bacterium.
Examples:
- EcoR I is isolated from Escherichia coli RY 13.
- Hind II is from Haemophilus influenza.
- Bam H I is from Bacillus amylotiquefaciens.
- Sal I is from Streptomyces Albus
- Pst I is from Providencia stuartii.
Question 4.
Explain any three methods of vector less gene transfer. (CBSE Outside Delhi 2013)
Answer:
Vectors of gene transfer. Following are common methods of vectors gene transfer.
- Microinjection: Microinjection is the process/technique of introducing foreign genes into a host cell by injecting the DNA directly into the nucleus by using microneedle or micropipette.
- Electroporation: Electroporation is the process by which transient holes are produced in the plasma membrane of the (host) cell to facilitate entry of foreign DNA.
- Gene Gun: Gene gun is the technique of bombarding microprojectiles (gold or tungsten particles) coated with foreign DNA with great velocity into the target cell.
Question 5.
Write a note on the cloning vector.
Answer:
Cloning vectors:
- Plasmids and bacteriophages are the commonly used vectors
- Presently genetically engineered/ synthetic vectors are also used for easily linking the foreign DNA and selection of recombinants from non-recombinants.
- The following features are required to facilitate cloning in a vector:
(a) Origin of replication (Ori)
(b) Selectable marker
(c) Cloning (Recognition) site
(d) Small size of the vector.
Question 6.
What is PCR? List the three main steps. Show the steps with a diagrammatic sketch.
Answer:
- PCR. Polymerase Chain Reaction.
- Three steps of PCR.
(a) Denaturation
(b) Primer annealing and
(c) Extension of primers.
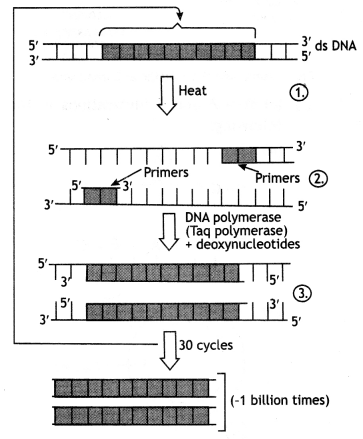
The three steps of PCR
Question 7.
Name the various cloning vectors and explain how a plasmid can be used for genetic engineering.
Answer:
Cloning vectors:
- Plasmids
- Bacteriophages
- Plant and animal vectors
- Jumping genes (Transposons)
- Artificial chromosomes of bacteria, yeast and mammals (BAC, YAC).
Use of plasmid as genetic material Plasmids are obtained from bacteria. They are treated with a restriction endonuclease enzyme to obtain the fragments of the desired genome. They are allowed to fuse with the help of a DNA ligase enzyme. The recombinant plasmids thus formed are used as genetic material.
Question 8.
Give various means by which a competent host is formed for recombinant DNA technology. Why and how bacteria can be made ‘competent’? (CBSE Delhi 2013)
Answer:
A host cell should be competent enough to take the DNA molecule for the transformation as the following methods can be used.
- Using divalent cations: Bacteria are treated with Ca2+, etc. so that DNA enters the bacterium through pores in its cell wall.
- Heat shock: Cells can be incubated on ice and then at 42°C for a heat shock and then again put on ice.
- Microinjection: Recombinant DNA is directly injected into the nucleus of an animal cell.
- Biolistic. Cells bombarded with high- velocity micro-particles of gold or tungsten coated with DNA is known as a gene gun.
Question 9.
How is recombinant DNA transferred to host?
Answer:
Transfer of recombinant DNA into the host:
- The bacterial cells must be made competent to take up DNA; this is done by treating them with a specific concentration of calcium, that increases the efficiency with which DNA enters the cell through the pores in its cell wall.
- Recombinant DNA can then be forced into such cells by incubating the cells with recombinant DNA on ice followed by placing them at 42°C and then putting them back on ice (heat shock treatment),
- Microinjection is a method in which the recombinant DNA is directly injected into the nucleus of the animal cell with the help of microneedles or micropipettes.
- Gene gun or biolistics is a method suitable for plant cells, where cells are bombarded with high-velocity microparticles of gold or tungsten coated with DNA.
- Disarmed pathogens are used as vectors; when they are allowed to infect the cell, they transfer the recombinant DNA into the host.
Question 10.
Why DNA cannot pass through the cell membrane? How can the bacteria be made competent to take up a plasmid? Explain a method for the introduction of alien DNA into a plant host cell. Name a pathogen that is used as a disarmed vector. (CBSE Outside Delhi 2019)
Answer:
DNA is a hydrophilic molecule thus it cannot pass through the cell membrane.
Bacterial cells are made ‘competent’ by treating them with a specific concentration of divalent cation such as calcium in order to take up the plasmid. The divalent cation increases the efficiency with which DNA enters the bacterium through the pores of the cell wall.
Procedure: Recombinant DNA is forced into ‘competent’ bacterial host cells by incubating them on the ice. It is followed by placing them briefly at 42°C. It is termed ‘heat stock’ treatment. Again they are placed back on ice. This process allows bacteria to take up the recombinant DNA.
Gene gun or biolistic method is used for the introduction of alien DNA into a plant host cell. Here, the plant cells are bombarded with high-velocity micro-particles of gold or tungsten coated with DNA. Agrobacterium tumefaciens or Retroviruses can be used as a disarmed vector.
Question 11.
Write a note on vectors used during recombinant DNA technology. (CBSE Delhi 2008)
Answer:
A vector or vehicle DNA is used as a carrier for transferring selected DNA into cells. A plasmid with its small DNA from a bacterium is a good choice for indirect gene transfer because it can move from one cell to another and make several copies of itself. However, artificial chromosomes from bacteria and yeast called BACs and YACs respectively are more efficient for eukaryotic gene transfers.
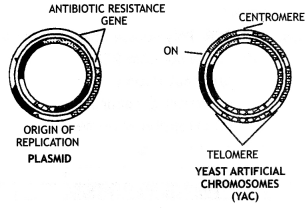
Plasmid and Yeast Artificial Chromosome
Question 12.
(i) Identify A and B illustrations in the following:
(a)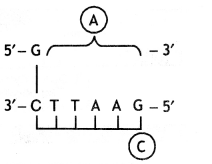
(b)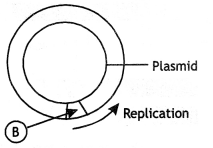
Answer:
A= 5′ GAATTC 3′
B marks for ORI (origin of replication).
(ii) Write the term given to A and C and why?
Answer:
A represents a nucleotide palindromic sequence. C-sticky end.
(iii) Expand PCR. Mention its importance in biotechnology. (CBSE Delhi 2011)
Answer:
PCR, Polymerase chain reaction. It helps in gene amplification.
Question 13.
Write the role of the following sites in pBR322 cloning vector:
(a) rop
Answer:
Role of rop, ori and selectable marker in pBR322 cloning vector.
Role of rop: Rop gene regulates copy number. Rop process is involved in stabilising the interaction between RNA I and RNA II which in turn prevents replication of pBR322.
(b) ori
Answer:
Origin of replication (Ori):
- It is a specific sequence of DNA bases, which is responsible for initiating replication.
- An alien DNA for replication should be linked to the origin of replication.
- A prokaryotic DNA has normally a single origin of replication, while eukaryotic DNA may have more than one origin of replication.
- The sequence is responsible for controlling the copy number of linked DNA.
(c) selectable marker (CBSE Delhi 2019 C)
Answer:
Selectable marker:
- A marker is a gene which helps in selecting those host cells which contain the vector (transformant) and eliminating the non-transformants.
- Common selectable markers for E. coli include the genes encoding resistance to antibiotics such as ampicillin. Chloramphenicol, tetracycline and kanamycin or the gene for B-galactosidase can be identified by a colour reaction.
Question 14.
(i) Explain the significance of palindromic nucleotide sequence in the formation of recombinant DNA.
Answer:
The palindromic sequences, i.e. the sequence of base pairs read the same on both the DNA strands when the orientation of reading is kept the same, e.g.
5’ — GAATTC — 3’
3’ — CTTAAG — 5’
Every endonuclease inspects the entire DNA sequence for palindromic recognition sequence.
(ii) Write the use of restriction endonuclease in the above process. (CBSE 2017)
Answer:
On finding the palindrome, the endonuclease binds to the DNA. It cuts the opposite strands of DNA, but between the same bases on both the strands and forms sticky ends. This sticky ends facilitate the action of enzyme DNA ligase and help in the formation of recombination DNA.
Question 15.
Describe the roles of heat, primers and the bacterium Thermus Aquaticus in the process of PCR. (CBSE 2017)
Answer:
Role of heat: Heat helps in the denaturation process in PCR. The double-stranded DNA is heated in this process at very high temperature (95°C) so that both the strands separate.
Role of primers: Primers are chemically synthesised small oligonucleotides of about 10-18 nucleotides. These are complementary to a region of template DNA and helps in the extension of the new chain. Rote of Bacterium Thermus
Aquaticus: A thermostable Taq DNA polymerase is isolated from this bacterium, which can tolerate high temperatures and forms new strand.
Question 16.
How has the use of Agrobacterium as vectors helped in controlling Meloidogyne incognita infestation in tobacco plants? Explain in the correct sequence. (CBSE 2018, Outside Delhi 2019)
Or
(a) Write the mechanism that enables Agrobacterium tumefaciens to develop tumours in their host dicot plant.
(b) State how Agrobacterium tumefaciens and some retroviruses have been modified as useful cloning vectors. (CBSE Delhi 2019 C)
Answer:
(a) Cloning
(b) A nematode Meloidogyne incognita infects the roots of tobacco plants and causes a great reduction in yield.
To prevent this infestation a novel strategy was adopted which was based on the process of RNA interference (RNAi).
Nematode-specific genes were introduced into the host plants using Agrobacterium vectors. The introduction of DNA was such that it produced both sense and anti-sense RNA in the host cells. These two RNAs, being complementary to each other, formed a double-stranded RNA (dsRNA) that initiated RNAi and thus, silenced specific mRNA of the nematode. Due to this the parasite could not survive in a transgenic host by expressing specific interfering RNA. The transgenic plant, therefore, got itself protected from the parasite.
Question 17.
Explain the roles of the following with the help of an example each in recombinant DNA technology:
(i) Restriction Enzymes
Answer:
Restriction enzymes :
(a) Restriction enzymes belong to nucleases class of enzymes which breaks nucleic acids by cleaving their phosphodiester bonds.
(b) Since restriction endonucleases cut DNA at a specific recognition site, they are used to cut the donor DNA to isolate the desired gene.
(c) The desired gene has sticky ends which can be easily ligated to cloning vector cut by same restriction enzymes having complementary sticky ends to form recombinant DNA.
(d) An example is EcoR1 which is obtained from E.coli bacteria “R” strain which cuts DNA at specific palindromic recognition site.
5‘ GAATTC 3‘
3‘ CTTAAG 5‘
(ii) Plasmids (CBSE 2018)
Answer:
Plasmids: Plasmids are autonomous, extrachromosomal circular double-stranded DNA of bacteria. They are used as cloning vectors in genetic engineering because they are small and self-replicating. Some plasmids have antibiotic resistance genes which can be used as marker genes to identify recombinant plasmids from non-recombinant ones.
To obtain the desired products, plasmids are cut and ligated with desired genes and transformed into a host cell for amplification. An example of artificially modified plasmids is pBR322 (constructed by Bolivar and Rodriguez) or pUC (constructed at University at California).
Question 18.
When the gene product is required in large amounts, the transformed bacteria with the plasmid inside the bacteria are cultured on a large scale in an industrial fermenter which then synthesises the desired protein. This product is extracted from the fermenter for commercial use.
(a) Why is the used medium drained out from one side while the fresh medium is added from the other? Explain.
Answer:
In the bioreactor used medium is drained out and the fresh medium is added to maintain the cells in their physiologically most active log / experimental phase,
(b) List any four optimum conditions for achieving the desired product in a bioreactor. (CBSE Sample Paper 2020)
Answer:
Condition for obtaining the desired product in a bioreactor:
- Temperature
- Substrates
- SaLts
- pH
- Oxygen
Question 19.
List the steps in the formation of rDNA.
Answer:
Steps in formation of rDNA:
Recombined DNA technology involves the following steps:
- Isolation of DNA.
- Fragmentation of DNA by restriction endonucleases.
- Isolation of the desired DNA fragment.
- Amplification of the gene of interest.
- Ligation of the DNA fragment into a vector using DNA ligase.
- Transfer of DNA fragment into the vector using DNA ligase.
Question 20.
How is the isolated gene of interest amplified? (CBSE Delhi 2019, 2019 C)
Answer:
Amplification of the DNA/gene of interest:
- Amplification refers to the process of making multiple copies of the DNA segment in vitro.
- It employs the polymerase chain reaction (PCR).
- The process was designed by K. Mullis,
- This technique involves three main steps:
(a) Denaturation
(b) Primer annealing and
(c) Extension of primers. - The double-stranded DNA is denatured by subjecting it to high temperatures.
- Two sets of primers are used; primers are the chemically synthesised short segments of DNA (oligonucleotides), that are complementary to the segment of DNA (of interest).
- DNA polymerase enzyme (Taq polymerase) is used to make copies of DNA making use of genomic template DNA and primer.
Question 21.
List the features required to facilitate cloning into a vector. Show with a sketch the E. coli cloning vector showing restriction sites.
Or
Sketch pBR322. (CBSE 2012, Outside Delhi 2019)
Answer:
Features required to facilitate cloning vector.
- Origin of replication (Ori)
- Selectable marker
- Cloning sites
- Vectors for cloning genes in plants and animals
Sites of cloning vector
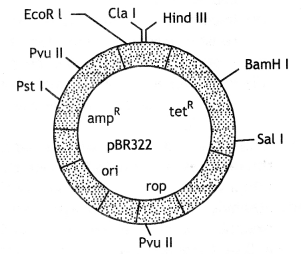
E. coli Cloning Vector pBr322 showing restriction sites (Hindlll, EcoRI, BamHI, Sal I, Pvu II, Pst I, ClaI), oriV and antibiotic resistance genes (ampR and tetR). Rop codes for the proteins involved in the replication of the plasmid.
Question 22.
With the help of simple sketch show the action of restriction enzyme (EcoR1).
Answer:
The action of restriction enzyme.
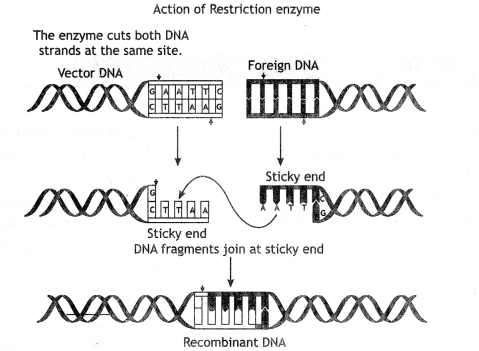
Ecol cuts the DNA between bases G and A only when the sequence GAATTC is present in the DNA.
Question 23.
Explain the importance of (a) ori, (b) ampR and (c) rop in the E. colt vector. (CBSE Outside Delhi 2009, Outside Delhi 2019)
Answer:
- Importance of ori: This is a sequence from where replication starts and any piece of DNA, when linked to this sequence, can be made to replicate within the host cells, it allows multiple copies per cell.
- Importance of ampR: It is the antibiotic resistance gene for ampicillin. It helps in the selection of transformer cells.
- Importance of rop: It codes for the proteins involved in the replication of plasmid.
Question 24.
Name any two cloning vectors. Describe the features required to facilitate cloning into a vector. (CBSE Sample Paper)
Answer:
Plasmids and bacteriophages are two examples of the cloning vector. A vector is a DNA molecule that has the ability to replicate in an appropriate host cell and into which the DNA fragment to be cloned is integrated for cloning.
A good vector must have the following properties:
- It should be able to replicate autonomously.
- it should be easy to isolate and purify,
- It should be easily introduced into the host cells.
Cloning vectors: All vectors that are used for propagation of DNA inserts in a suitable host are called cloning vectors. When a vector is designed for the expression of, i.e. production of the protein specified by, the DNA insert, it is termed as an expression vector.
Question 25.
What are bioreactors? Sketch the two types of bioreactors. What is the utility? Which is the common type of bioreactors? (CBSE Delhi 2013)
Or
How do bioreactors help in the production of recombinant proteins? (CBSE Outside Delhi 2009)
Or
(i) How has the development of bioreactor helped in biotechnology?
(ii) Name the most commonly used bioreactor and describe its working. (CBSE Delhi 2018, 2019 C)
Answer:
Small volume cultures cannot yield appreciable quantities of products. To produce these products in large quantities the development of ‘bioreactors’ was required where large volumes (100-1000 litres) of culture can be processed. Thus bioreactors can be thought of as vessels in which raw materials are biologically converted into specific products, using microbial, plant, animal or human cells or individual enzymes.
Role. A bioreactor provides the optimal conditions for achieving the desired product by providing optimum growth conditions (temperature, pH, substrate, salts, vitamins, oxygen).
One of the most commonly used bioreactors is of stirring type.
A stirred tank reactor is cylindrical or a container with a curved base which facilitate the mixing of the reactor contents. The stirrer facilitates even mixing and oxygen availability throughout the bioreactor. Alternatively, air can be passed through the reactor. It consists of agitator system, an oxygen delivery system, a foam control system, a temperature control system, pH control system and sampling ports so that small volumes of the culture can be withdrawn periodically.
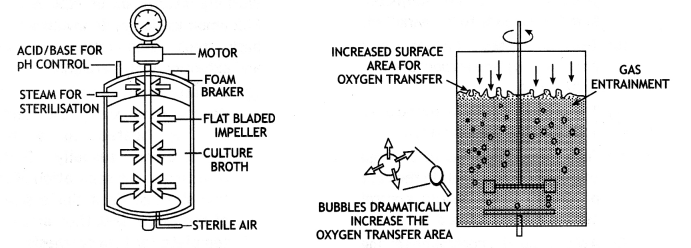
(a) Simple stirred-tank bioreactor (b) Sparged stirred-tank bioreactor through which sterile air bubbles are sparged
Question 26.
Describe briefly the following:
(i) Origin of replication (Ori).
Answer:
(a) It is a specific sequence of DNA bases, which is responsible for initiating replication.
(b) An alien DNA for replication should be linked to the origin of replication.
(c) A prokaryotic DNA has normally a single origin of replication, while eukaryotic DNA may have more than one origin of replication.
(d) The sequence is responsible for controlling the copy number of linked DNA.
(ii) Bioreactor.
Answer:
(a) They are vessels in which raw materials are biologically converted into specific products using microbial, plant or human cells.
(b) A bioreactor provides optimal conditions for achieving the desired product by providing optimum growth conditions, pH, substrate salts, vitamins, oxygen, etc.
(c) The commonly used bioreactors are of stirring type.
(d) A stirred-tank reactor is usually cylindrical or with a curved base to facilitate the mixing of the contents.
(e) The stirrer facilitates the even-mixing and oxygen availability throughout the bioreactor.
(f) The bioreactor has the following components:
- An agitator system.
- An oxygen delivery system.
- A foam control system.
- A temperature control system.
- pH control system and
- Sampling ports.
(iii) Downstream processing.
Answer:
(a) It refers to the series of processes, to which a genetically modified product has to be subjected before it is ready to be marketed.
(b) The processes include two processes:
- separation and
- purification.
(c) The product has to be formulated with suitable preservatives.
(d) Such formulation has to undergo thorough clinical trials in the case of drugs. Strict quality control testing is also required.
(e) A proper quality controlled testing of each product is also required.
Question 27.
Besides better aeration and mixing properties, what other advantages do stirred tank bioreactors to have over shake flasks?
Answer:
Shake flasks are the conventional flasks for fermentation studies during secondary screening or laboratory process development. So, stirred-tank bioreactors are used to produce the product in large quantities.
Besides aeration and mixing:
- it also helps in providing optimum growth conditions (temperature, pH, substrate, salts, vitamins, oxygen) to achieve the desired product.
- cost-effective
- due to baffles, the oxygen transfer rate is very high
- the capacity of fermenters is more.
Question 28.
Explain briefly the following
(i) PCR
(ii) Restriction enzymes and DNA
(iii) Chitinase. (CBSE 2012)
Or
Explain the three steps involved in a polymerase chain reaction. (CBSE Delhi 2018C)
Answer:
(i) PCR-Polymerase Chain Reaction; It is the process in which multiple copies of the gene or segment of DNA of interest are synthesised in vitro using primers and DNA polymerase.
Working Mechanism of PCR: A single PCR amplification cycle involves three basic steps: denaturation, annealing and extension (polymerisation).
(a) Denaturation. In the denaturation step, the target DNA is heated to a high temperature (usually 94°C), resulting in the separation of the two strands. Every single strand of the target DNA then acts as a template for DNA synthesis.
(b) Annealing (Anneal = Join). In this step, the two oligonucleotide primers anneal (hybridize) to each of the single-stranded template DNA since the sequence of the primers is complementary to the 3’ ends of the template DNA. This step is carried out at a lower temperature depending on the length and sequence of the primers.
(c) Primer Extension (Polymerisation): The final step is an extension, wherein Taq DNA polymerase (of a thermophilic bacterium Thermus aquatics) causes synthesis of the DNA region between the primers, using dNTPs (deoxynucleoside triphosphates) and Mg2+. It means the primers are extended towards each other so that the DNA segment lying between the two primers is copied.
The optimum temperature for this polymerisation step is 72°C. To begin the second cycle, the DNA is again heated to convert all the newly synthesised DNA into single strands, each of which can now serve as a template for synthesis of more new DNA. Thus the extension product of one cycle can serve as a template for subsequent cycles and each cycle essentially doubles the amount of DNA from the previous cycle. As a result, from a single template molecule, it is possible to generate 2n molecules after n number of cycles.
Applications of PCR:
- Diagnosis of pathogen
- Diagnosis of the specific mutation
- DNA fingerprinting
- Detection of plant pathogens
- Cloning of DNA fragments from mummified remains of humans and extinct animals.
(ii) Restriction enzymes:
(a) They are called “molecular scissors” or chemical scalpels.
(b) Restriction enzymes, synthesised by micro-organisms as a defence mechanism, are specific endonucleases, which can cleave double-stranded DNA.
(c) Restriction enzymes belong to a class of enzymes called nucleases.
(d) They are of two kinds:
- Exonucleases, which remove nucleotides from the ends of DNA.
- Endonucleases, which cut the DNA at specific positions anywhere in its length (within).
(e) The recognition sequence is a palindrome, where the sequence of base pairs reads the same on both the DNA strands when the orientation of reading is kept the same, i.e. 5′ → 3′ direction or 3′ → 5′ direction.
e.g. 5′ – GAATTC – 3′
3′ – CTTAAG – 5′
(f) Each restriction endonuclease functions by inspecting the length of a DNA sequence and binds to the DNA at the recognition sequence.
(g) It cuts the two strands of the double helix at specific points in their sugar-phosphate backbones, a little away from the centre of the palindrome sites, but between the same two bases on both the strands.
(h) As a result, single-stranded portions called sticky ends are produced at the ends of the DNA; this stickiness of the end facilitates the action of enzyme DNA ligase.
- When cut by the same restriction endonuclease, the DNA fragments (of the donor as well as the host/ recipient) yield the same kind of ‘sticky ends’ which can be joined end-to-end by DNA ligases.
- Chitinase. This cell wall in fungi is made of chitin. The enzyme is used in fungi to break open the cell to release DNA along with their macromolecules like RNA proteins, lipids and polysaccharides.
Question 29.
Discuss with your teacher and find out how to distinguish between
(i) Plasmid DNA and chromosomal DNA
Answer:
Differences between plasmid DNA and chromosomal DNA:
| Plasmid DNA | Chromosomal DNA |
| 1. It is self-replicating, DNA molecule found naturally in many bacteria and yeast. | 1. Chromosomal DNA present in chromosomes of all organisms. |
| 2. It is not essential for normal growth and division. | 2. It Is essential for growth and division. |
| 3. It contains information for a few traits. | 3. It contains information for all traits. |
(ii) Exonuclease and Endonuclease (CBSE, Delhi 2013)
Answer:
Differences between Exonuclease and Endonuclease:
| Endonuclease | Exonuclease |
| It cuts the DNA at a specific position of nitrogen bases anywhere within the length of DNA except the ends. | This enzyme removes nucleotides from the terminals from 5’ or 3’ ends of DNA molecules. |
Question 30.
Collect the examples of palindromic sequences by consulting your teacher. Better try to create a palindromic sequence by following base pair rules.
Answer:

Question 31.
Can you list 10 recombinant proteins which are used In medical practice? Find where they are used as therapeutics (use the Internet).
Answer:
| Recombinant Protein | Therapeutic Use |
| 1. Insulin | For the treatment of diabetes Mellitus |
| 2. Human Growth Hormone | For the treatment of dwarfism |
| 3. Interferons | For the treatment of viral diseases, cancer and AIDS. |
| 4. Streptokinase | For treating thrombosis. |
| 5. Tumour Necrosis factor | For treating sepsis and cancer |
| 6. Interleukins | For treating various cancers. |
| 7. Hepatitis-B Surface Antigen | The vaccine against Hepatitis- B |
| 8. Granulocyte Colony-stimulating factor | For treating cancer and AIDS and in bone marrow transplantation |
| 9. Granulocyte-macrophage Colony-stimulating factor | For treating cancer and AIDS |
| 10. Bovine growth hormone | For increasing milk yield. |
Question 32.
How is DNA isolated in a purified form? (CBSE Outside Delhi 2009)
Answer:
Isolation of DNA in the purified form:
- DNA has to be isolated in pure form for the action of restriction enzymes.
- DNA can be released from the cells by digesting the cell envelope by the use of enzymes like lysozyme for bacterial cells, chitinase for fungal cells and cellulase for plants cells.
- Since DNA is intertwined with histone proteins and RNAs, proteins are removed by treatment with proteases and RNAs by ribonucleases.
- Other impurities are removed by employing suitable treatments.
- The purified DNA is precipitated by the addition of chilled ethanol. It is seen as fine threads in suspension.
Question 33.
How is isolation and Fragmentation of DNA of interest carried out in recombinant DNA technology? (CBSE Outside Delhi 2009, 2019)
Answer:
1. Fragmentation DNA: Fragmentation of DNA is carried out by incubating the purified DNA molecules with suitable restriction enzymes at optimal conditions of temperature and pH.
2. Isolation of DNA (gene) of Interest:
(a) The fragments of DNA are separated by a technique called gel electrophoresis.
(b) The DNA is cut into fragments by restriction endonucleases.
(c) These fragments are separated by a technique called gel electrophoresis.
(d) Agarose, the natural polymer obtained from seaweeds, is used as the matrix.
(e) DNA fragments being negatively charged are separated by forcing them to move through the matrix towards the anode under an electric field.
(f) The DNA fragments separate/ resolve according to their size.
(g) The separated molecules are stained by ethidium bromide and visualised by exposure to UV radiation, as bright orange coloured bands.
(h) The separated bands of DNA (on the gel) are cut from the gel and extracted from the gel piece (elution).
(i) Such DNA fragments are purified and used for constructing recombinant DNA by joining them with cloning vectors.